-
PDF
- Split View
-
Views
-
Cite
Cite
Fabrice Roux, Maria Colomé-Tatché, Cécile Edelist, René Wardenaar, Philippe Guerche, Frédéric Hospital, Vincent Colot, Ritsert C Jansen, Frank Johannes, Genome-Wide Epigenetic Perturbation Jump-Starts Patterns of Heritable Variation Found in Nature, Genetics, Volume 188, Issue 4, 1 August 2011, Pages 1015–1017, https://doi.org/10.1534/genetics.111.128744
Close - Share Icon Share
Abstract
We extensively phenotyped 6000 Arabidopsis plants with experimentally perturbed DNA methylomes as well as a diverse panel of natural accessions in a common garden. We found that alterations in DNA methylation not only caused heritable phenotypic diversity but also produced heritability patterns closely resembling those of the natural accessions. Our findings indicate that epigenetically induced and naturally occurring variation in complex traits share part of their polygenic architecture and may offer complementary adaptation routes in ecological settings.
THE production of new heritable phenotypic variation is an essential aspect of evolution. It shapes the capacity of a population to adapt to environmental change and thus provides the raw material for natural selection. Our current view posits rare DNA sequence mutations as the primary source of this process (Ossowski et al. 2010). In plants and animals, drastic changes in DNA methylation may provide a complementary route to novel heritable variants. Transient disruption of the DNA methylation maintenance machinery in the flowering plant Arabidopsis thaliana, for example, triggers the repatterning of DNA methylation states at thousands of loci and leads to the remobilization of some transposable elements (Vongs et al. 1993; Johannes et al. 2009; Mirouze et al. 2009; Reinders et al. 2009; Teixeira et al. 2009; Tsukahara et al. 2009). These rapid changes cause phenotypic effects that persist over many generations even upon outcrossing to wild-type plants (Johannes et al. 2009; Reinders et al. 2009). Here we document such effects for a spectrum of complex traits relevant for adaptation and show that they produce heritability patterns resembling those found in natural populations that have undergone thousands of years of divergent evolution.
We examined a large panel of Arabidopsis epigenetic recombinant inbred lines (epiRILs) (Johannes et al. 2009) under ecologically realistic conditions. This population was derived from a cross between two parents with nearly identical genomes but highly divergent epigenomes as a result of a mutation (in one of the parents) in DDM1, a gene essential for DNA methylation and the silencing of repeat elements and some genes (Vongs et al. 1993). After backrossing of the F1 and selection of the progeny homozygous for wild-type DDM1, the epiRILs were propagated through six rounds of selfing and were found to segregate many of the epigenetic as well as a few nucleotide changes harbored by the ddm1 parent (Johannes et al. 2009; Teixeira et al. 2009). The epiRILs therefore permit a detailed assessment of the long-term molecular and phenotypic consequences of these stable changes (Johannes et al. 2008; Johannes and Colomé-Tatché 2011).
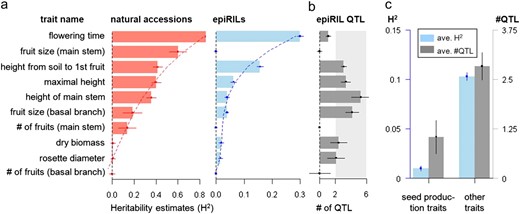
We grew 5724 epiRILs (477 lines × 12 replicates) along with their two parental lines (2 × 120 replicates) in a common garden (supporting information, Figure S2). For direct comparison, we also grew a collection of Arabidopsis natural accessions (10 ecotypes × 36 replicates). These accessions (An-1, En-1, Gr-1, Hs-0, Is-0, Jm-0, Ka-0, Nd-1, Per-1, and Sap-0) were chosen to sample the worldwide phenotypic diversity of this species (File S1). All plants (n ≈ 6300) were profiled for up to 10 complex traits (Figure 1a and File S1).
(a) Heritability estimates (±SE) for 10 traits in the natural accessions and epiRILs with weighted least-square cubic fit. (b) Estimates of the number of QTL (±SE) in the epiRILs. The light gray rectangle indicates a polygenic architecture with a theoretical upper limit of 6 QTL. (c) Average estimates of heritability and QTL number contrasted between seed production traits (fruit size and fruit number) and all other traits.
We found significant broad-sense heritability (H2) in the epiRILs for most traits (estimated from variance component analysis, see File S1). H2 ranged from 4% for fruit size on primary branches to 30% for flowering time (Figure 1a and Table S2). These estimates were consistent with a predicted polygenic architecture involving on the order of two to five quantitative trait loci (QTL) (Johannes and Colomé-Tatché 2011); the only exception was flowering time, for which a single-locus inheritance model could still not be ruled out (Figure 1b and File S1). In comparison to both of the parental lines, the phenotypic means of the epiRILs were closer to those of the wild type (Figure S1 and Table S1). This agrees with the crossing scheme used to derive the epiRILs (backcross to wild-type base population) (Johannes et al. 2009; Johannes and Colomé-Tatché 2011) as well as with the progressive RNA-directed remethylation of specific DNA sequences previously documented in this system (Teixeira et al. 2009) and thus further suggests that the QTL underlying heritability originate from the parental generation rather than from a later stage of inbreeding (Johannes and Colomé-Tatché 2011).
To understand the epigenetically induced heritable effects observed in the epiRILs in the wider context of natural variation, we compared the epiRILs directly with the panel of natural accessions grown in the same environment. Overall, the accessions revealed higher heritability for most phenotypes (Figure 1a and Table S2), which was perhaps expected on the basis of their diverse geographical origins (see File S1). However, when we examined the distribution of heritability values across all traits, we found striking parallels between the epiRILs and natural accessions (Figure 1a and Table S2). This observation is remarkable given that the latter have likely evolved independently from each other for thousands of years, whereas the epiRILs are the product of a single epigenomic perturbation event in a common founder eight generations earlier. A parsimonious explanation is that many of the QTL segregating in the epiRILs act through a heritable architecture that is common to both populations. This possibility is supported by the observation that the epiRILs and the accessions show significantly similar “genetic” correlations across all of the 10 traits (Mantel test: P = 0.0013). Whether the epiRILs QTL physically overlap with those previously mapped in Arabidopsis natural populations (Atwell et al. 2010), or present new entry points in a common regulatory network, remains to be seen.
Specific exceptions to the similarities between the epiRILs and the natural accessions were the relatively low heritability values in the epiRILs for seed production traits (i.e., the number and size of fruits) compared to all other traits (Figure 1, a and c, and Table S2). One hypothesis is that the initial epigenetic changes in the mutant parent did not affect loci involved in the control of these traits. This explanation predicts similar phenotypic means for the wild-type and mutant parents, which was clearly not the case (see Figure S1 and Table S1). The causative variants (or the phenotypic effects of these variants) therefore appear to have been lost at some later time during inbreeding. We find that this is unlikely the result of selection, to the extent that only 5 of 509 lines (0.8%) failed during epiRIL construction (Johannes et al. 2009). Hence, other mechanisms such as phenotypic buffering (Fu et al. 2009) or locus-specific epigenetic editing may be responsible. Irrespective of the underlying processes, the fact that heritable variation appears in this trait-specific manner in the epiRILs suggests that plants may have evolved mechanisms for exploiting epigenomic perturbation events that bypass the adverse effects of inbreeding depression.
In summary, our findings clearly demonstrate that transient perturbations of epigenetic systems can rapidly generate heritable variation for complex traits that is similar to that observed between divergent natural populations. Whether causative heritable variants produced in this way are the result of stable epigenetic changes (epimutations) or DNA sequence mutations, such as those associated with mobilization of transposable elements, remains to be determined experimentally. Regardless of their physical basis, these heritable variants can become ready targets of natural or artificial selection and shape the evolutionary trajectory of the species.
Acknowledgements
We thank two reviewers for their helpful comments. We appreciate the input from O. Loudet, T. Mackay, M. Nordborg, and B. Walsh on earlier versions of this manuscript. We thank N. Faure and J. Conynck for help in phenotyping. We acknowledge support from Genoplante (to P.G., F.H., and V.C.), the Netherlands Organisation for Scientific Research (NWO) (to F.J. and M.C.-T.), the Netherlands Bioinformatics Centre (NBIC) (to R.W.).
Footnotes
Communicating editor: J. Borevitz
Literature Cited
Author notes
These authors contributed equally to this work.
Supporting information is available online at http://www.genetics.org/content/suppl/2011/05/19/genetics.111.128744.DC1.