-
PDF
- Split View
-
Views
-
Cite
Cite
Jian-Qiu Wu, Yanfang Ye, Ning Wang, Thomas D Pollard, John R Pringle, Cooperation Between the Septins and the Actomyosin Ring and Role of a Cell-Integrity Pathway During Cell Division in Fission Yeast, Genetics, Volume 186, Issue 3, 1 November 2010, Pages 897–915, https://doi.org/10.1534/genetics.110.119842
Close - Share Icon Share
Abstract
A major question about cytokinesis concerns the role of the septin proteins, which localize to the division site in all animal and fungal cells but are essential for cytokinesis only in some cell types. For example, in Schizosaccharomyces pombe, four septins localize to the division site, but deletion of the four genes produces only a modest delay in cell separation. To ask if the S. pombe septins function redundantly in cytokinesis, we conducted a synthetic-lethal screen in a septin-deficient strain and identified seven mutations. One mutation affects Cdc4, a myosin light chain that is an essential component of the cytokinetic actomyosin ring. Five others cause frequent cell lysis during cell separation and map to two loci. These mutations and their dosage suppressors define a signaling pathway (including Rho1 and a novel arrestin) for repairing cell-wall damage. The seventh mutation affects the poorly understood RNA-binding protein Scw1 and severely delays cell separation when combined either with a septin mutation or with a mutation affecting the septin-interacting, anillin-like protein Mid2, suggesting that Scw1 functions in a pathway parallel to that of the septins. Taken together, our results suggest that the S. pombe septins participate redundantly in one or more pathways that cooperate with the actomyosin ring during cytokinesis and that a septin defect causes septum defects that can be repaired effectively only when the cell-integrity pathway is intact.
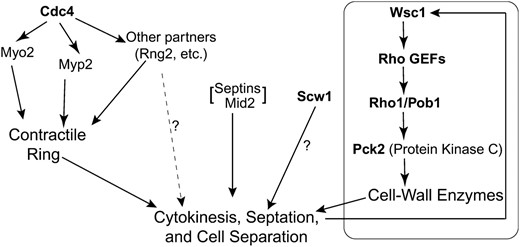
THE fission yeast Schizosaccharomyces pombe provides an outstanding model system for studies of cytokinesis (McCollum and Gould 2001; Balasubramanian et al. 2004; Pollard and Wu 2010). As in most animal cells, successful cytokinesis in S. pombe requires an actomyosin ring (AMR). The AMR begins to assemble at the G2/M transition and involves the type II myosin heavy chains Myo2 and Myp2 and the light chains Cdc4 and Rlc1 (Wu et al. 2003). Myo2 and Cdc4 are essential for cytokinesis under all known conditions, Rlc1 is important at all temperatures but essential only at low temperatures, and Myp2 is essential only under stress conditions. As the AMR constricts, a septum of cell wall is formed between the daughter cells. The primary septum is sandwiched by secondary septa and subsequently digested to allow cell separation (Humbel et al. 2001; Sipiczki 2007). Because of the internal turgor pressure of the cells, the proper assembly and structural integrity of the septal layers are essential for cell survival.
Septum formation involves the β-glucan synthases Bgs1/Cps1/Drc1, Bgs3, and Bgs4 (Ishiguro et al. 1997; Le Goff et al. 1999; Liu et al. 1999, 2002; Martín et al. 2003; Cortés et al. 2005) and the α-glucan synthase Ags1/Mok1 (Hochstenbach et al. 1998; Katayama et al. 1999). These synthases are regulated by the Rho GTPases Rho1 and Rho2 and the protein kinase C isoforms Pck1 and Pck2 (Arellano et al. 1996, 1997, 1999; Nakano et al. 1997; Hirata et al. 1998; Calonge et al. 2000; Sayers et al. 2000; Ma et al. 2006; Barba et al. 2008; García et al. 2009b). The Rho GTPases themselves appear to be regulated by both GTPase-activating proteins (GAPs) and guanine-nucleotide-exchange factors (GEFs) (Nakano et al. 2001; Calonge et al. 2003; Iwaki et al. 2003; Tajadura et al. 2004; Morrell-Falvey et al. 2005; Mutoh et al. 2005; García et al. 2006, 2009a,b). In addition, septum formation and AMR function appear to be interdependent. In the absence of a normal AMR, cells form aberrant septa and/or deposit septal materials at random locations, whereas a mutant defective in septum formation (bgs1) is also defective in AMR constriction (Gould and Simanis 1997; Le Goff et al. 1999; Liu et al. 1999, 2000). Both AMR constriction and septum formation also depend on the septation initiation network involving the small GTPase Spg1 (McCollum and Gould 2001; Krapp and Simanis 2008). Despite this considerable progress, many questions remain about the mechanisms and regulation of septum formation and its relationships to the function of the AMR.
One major question concerns the role(s) of the septins. Proteins of this family are ubiquitous in fungal and animal cells and typically localize to the cell cortex, where they appear to serve as scaffolds and diffusion barriers for other proteins that participate in a wide variety of cellular processes (Longtine et al. 1996; Gladfelter et al. 2001; Hall et al. 2008; Caudron and Barral 2009). Despite the recent progress in elucidating the mechanisms of septin assembly (John et al. 2007; Sirajuddin et al. 2007; Bertin et al. 2008; McMurray and Thorner 2008), the details of septin function remain obscure. However, one prominent role of the septins and associated proteins is in cytokinesis. Septins concentrate at the division site in every cell type that has been examined, and in Saccharomyces cerevisiae (Hartwell 1971; Longtine et al. 1996; Lippincott et al. 2001; Dobbelaere and Barral 2004) and at least some Drosophila (Neufeld and Rubin 1994; Adam et al. 2000) and mammalian (Kinoshita et al. 1997; Surka et al. 2002) cell types, the septins are essential for cytokinesis. In S. cerevisiae, the septins are required for formation of the AMR (Bi et al. 1998; Lippincott and Li 1998). However, this cannot be their only role, because the AMR itself is not essential for cytokinesis in this organism (Bi et al. 1998; Korinek et al. 2000; Schmidt et al. 2002). Moreover, there is no evidence that the septins are necessary for AMR formation or function in any other organism. A further complication is that in some cell types, including most Caenorhabditis elegans cells (Nguyen et al. 2000; Maddox et al. 2007) and some Drosophila cells (Adam et al. 2000; Field et al. 2008), the septins do not appear to be essential for cytokinesis even though they localize to the division site.
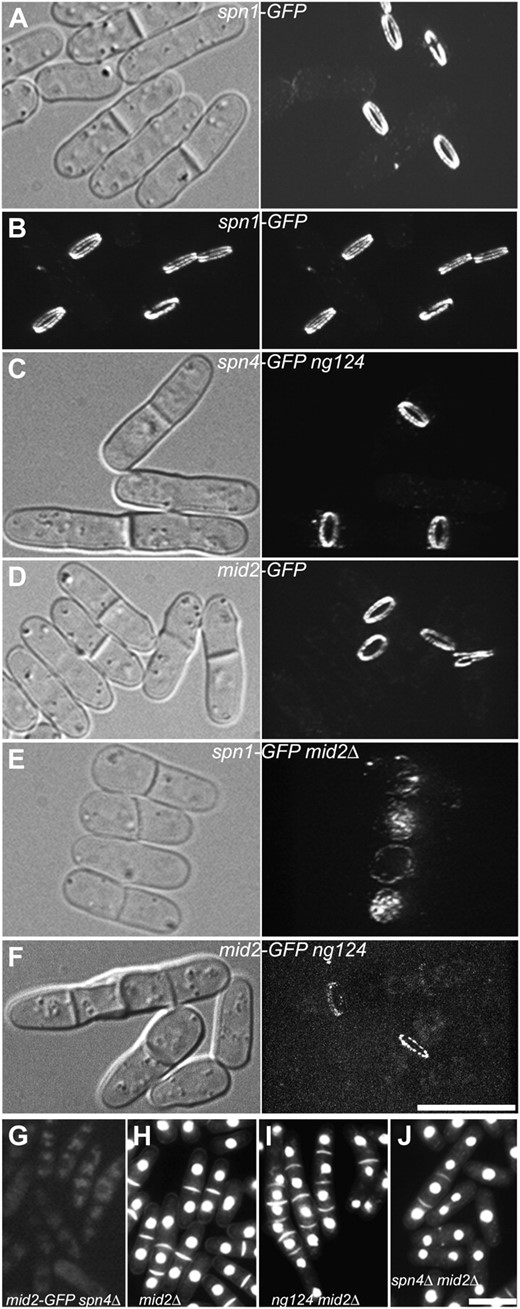
S. pombe has seven septins, four of which (Spn1, Spn2, Spn3, and Spn4) are expressed in vegetative cells and localize to the division site shortly before AMR constriction and septum formation (Longtine et al. 1996; Berlin et al. 2003; Tasto et al. 2003; Wu et al. 2003; An et al. 2004; Petit et al. 2005; Pan et al. 2007; Onishi et al. 2010). Spn1 and Spn4 appear to be the core members of the septin complex (An et al. 2004; McMurray and Thorner 2008), and mutants lacking either of these proteins do not assemble the others at the division site. Assembly of a normal septin ring also depends on the anillin-like protein Mid2, which colocalizes with the septins (Berlin et al. 2003; Tasto et al. 2003). Surprisingly, mutants lacking the septins are viable and form seemingly complete septa with approximately normal timing. These mutants do, however, display a variable delay in separation of the daughter cells, suggesting that the septins play some role(s) in the proper completion of the septum or in subsequent processes necessary for cell separation (Longtine et al. 1996; An et al. 2004; Martín-Cuadrado et al. 2005).
It is possible that the septins localize to the division site and yet are nonessential for division in some cell types because their role is redundant with that of some other protein(s) or pathway(s). To explore this possibility in S. pombe, we screened for mutations that were lethal in combination with a lack of septins. The results suggest that the septins cooperate with the AMR during cytokinesis and that, in the absence of septin function, the septum is not formed properly, so that an intact system for recognizing and repairing cell-wall damage becomes critical for cell survival.
MATERIALS AND METHODS
Strains, plasmids, growth conditions, and genetic methods:
S. pombe strains are listed in Table 1 and/or described below; all are congenic to strain 972 (Leupold 1970). Plasmid pREP42∷GFP-cdc4 was described by Balasubramanian et al. (1997). Plasmids with overlapping inserts covering nucleotides 627,249–681,978 of chromosome III were provided by the Yeast Genetic Resource Center Japan (YGRC/NBRP; http://yeast.lab.nig.ac.jp/nig/). Three S. pombe genomic DNA libraries were used: one with large inserts (provided by P. Young, Queens University, Ontario, Canada), constructed in plasmid pWH5 (Wright et al. 1986), and two with smaller inserts (pURSP1 and pURSP2, provided by A. Carr, University of Sussex, Falmer, UK), constructed in plasmid pUR19.
S. pombe strains used in this study
Strain . | Genotypea . | Source/reference . |
|---|---|---|
| 972 | h− wild type | Leupold (1970) |
| DM1274 | h− scw1Δ∷ura4+ ade6-M210 leu1-32 ura4-D18 | Jin and McCollum (2003) |
| FY435 | h+ ade6-M210 his7-366 leu1-32 ura4-D18 | S. Forsburg |
| FY11065 | h− cds1-2HA-6His∷ura4+ leu1 ura4 | Yeast Genetic Resource Center Japan |
| KS3320 | h+ swi5Δ∷kanMX6 | Anders et al. (2008) |
| KS3250 | h− rec12Δ∷kanMX6 ade6-M210 his7-366 leu1-32 lys1-37 | Anders et al. (2008) |
| MBY580 | h− bgs1-191 ade6-M210 leu1-32 lys1-131 ura4-D18 | Liu et al. (1999)b |
| TP5 | h− myp2Δ∷his7+ ade6-M210 his7-366 leu1-32 ura4-D18 | Bezanilla et al. (1997) |
| TP90 | h+ myo2-E1 myp2-Δ1∷his7+ade6-M216 his7-366 leu1-32 ura4-D18 | M. Bezanilla and T. Pollard |
| YDM74 | h− myo2-E1 ade6 his3-D1 leu1-32 ura4-D18 | D. McCollumc |
| JW21 | h+ cdc4-8 | P. Nurse; Nurse et al. (1976); McCollum et al. (1995) |
| JW81 | h− ade6-M210 leu1-32 ura4-D18 | Lab collection |
| JW182 | h− spn4-Δ2 | This studyd |
| JW183 | h− spn4+-GFP | This studyd |
| JW215 | h+ his3-27 leu1-32 ura4-D18 | Lab collection |
| JW249 | h− spn1-Δ2 | This studyd |
| JW251 | h− spn1+-GFP | This studyd |
| JW267 | h− 81nmt1-spn1+ 81nmt1-spn4+ leu1-32 ura4-D18 | See text. |
| JW289 | h+ spn1-Δ2 leu1-32 ura4-D18 | JW215 × JW249 |
| JW290 | h− spn1-Δ2 his3-27 ura4-D18 | JW215 × JW249 |
| JW295 | h+ spn4-Δ2 leu1-32 ura4-D18 | JW215 × JW182 |
| JW306 | h+ spn1+-GFP his3-27 leu1-32 ura4-D18 | JW215 × JW251 |
| JW314 | spn4-Δ2 mid2Δ∷ura4+ leu1-32 ura4-D18 | JW295 × JW430 |
| JW315 | scw1-ng124 mid2Δ∷ura4+ leu1-32 ura4-D18 | ng124e × JW430 |
| JW318 | spn1+-GFP mid2Δ∷ura4+ leu1-32 ura4-D18 | JW306 × JW430 |
| JW320 | scw1-ng124 spn4+-GFP leu1-32 ura4-D18 | JW331 × JW183 |
| JW321-1 | scw1-ng124 spn1-Δ2 ura4-D18 | JW331 × JW290 |
| JW326 | h− mid2+-GFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW330 | h− scw1-ng124 ade6-M210 leu1-32 ura4-D18 | This studyf |
| JW331 | h+ scw1-ng124 ade6-M210 leu1-32 ura4-D18 | This studyf |
| JW332 | mid2+-GFP spn4-Δ2 leu1-32 ura4-D18 | JW326 × JW295 |
| JW345 | cdc4-s16 myp2Δ∷his7+ ade6-M210 his7-366 leu1-32 ura4-D18 | JW400 × TP5 |
| JW374 | art1-s34 spn1-Δ2 leu1-32 ura4-D18 | JW403 × JW289 |
| JW380 | h− myp2Δ∷his7+ spn1-Δ2 ade6 his7-366 leu1-32 ura4-D18 | This studyg |
| JW396 | h− s26 leu1-32 ura4-D18 | This studyh |
| JW397 | h− s63 his3-27 leu1-32 ura4-D18 | This studyh |
| JW398 | h+ s28 leu1-32 ura4-D18 | This studyh |
| JW399 | h− cdc4-s16 leu1-32 ura4-D18 | This studye |
| JW400 | h+ cdc4-s16 ade6-M210 his7-366 leu1-32 ura4-D18 | JW399 × FY435 |
| JW401 | cdc4-s16 spn4-Δ2 leu1-32 ura4-D18 | JW399 × JW295 |
| JW402 | h− art1-s34 81nmt1-spn1+ 81nmt1-spn4+ leu1-32 ura4-D18 | This studyi |
| JW403 | h− art1-s34 leu1-32 ura4-D18 | This studye |
| JW404 | h+ art1-s34 leu1-32 ura4-D18 | This studye |
| JW405 | h− rgf3-s44 81nmt1-spn1+ 81nmt1-spn4+ leu1-32 ura4-D18 | This studyi |
| JW406 | h− rgf3-s44 his3-27 leu1-32 ura4-D18 | This studye |
| JW407 | h+ rgf3-s44 his3-27 leu1-32 ura4-D18 | This studye |
| JW409 | h− rgf1-GFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW412 | h− rgf3-GFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW423 | h− rgf1-Δ1 ade6 leu1-32 ura4-D18 | This studyj |
| JW430 | h− mid2Δ∷ura4+ade6 leu1-32 ura4-D18 | D. Brunner and P. Nurse |
| JW729 | h+ ade6-M210 leu1-32 ura4-D18 | Lab collection |
| JW903 | h+ 3nmt1-rgf2 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1078 | h− 3nmt1-rgf1 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1081 | h− 3nmt1-rgf3 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1084 | h− sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1100 | h+ spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1105 | h+ rgf3-mEGFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1113 | h− spn1-mEGFP sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | JW1100 × JW1084 |
| JW1122 | 3nmt1-rgf3 spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | JW1081 × JW1100 |
| JW1123 | 3nmt1-rgf1 spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | JW1078 × JW1100 |
| JW1124 | rgf1-GFP sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | This studyk |
| JW1125 | rgf3-s44 spn1-mEGFP leu1-32 ura4-D18 | JW406 × JW1100 |
| JW1126 | art1-s34 spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | JW403 × JW1100 |
| JW1128 | rgf3-GFP spn1-Δ2 ade6-M210 leu1-32 ura4-D18 | JW412 × JW289 |
| JW1131 | rgf3-mEGFP sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | JW1105 × JW1084 |
| JW1139 | rgf1-GFP spn1-Δ2 ade6-M210 leu1-32 ura4-D18 | JW409 × JW289 |
| JW1203 | mid2-GFP scw1-ng124 ade6-M210 leu1-32 ura4-D18 | JW326 × JW331 |
| JW2068 | h+ sec15Δ∷kanMX4 ade6 leu1-32 ura4-D18 | Lab collection |
| JW2155 | h− scw1-ng124 ade6-M210 leu1-32 ura4-D18 | This studyl |
| JW2271 | h+ scw1-ng124 ade6-M216 leu1-32 ura4-D18 | This studyl |
| JW2286 | scw1Δ∷ura4+ spn1-Δ2 ade6-M210 leu1-32 ura4-D18 | DM1274 × JW289 |
| JW2499 | cdc4-s16 scw1-ng124 ade6 leu1-32 ura4-D18 | JW399 × JW2271 |
| JW2503 | myo2-E1 scw1-ng124 ade6 leu1-32 ura4-D18 | YDM74 × JW2271 |
| JW2560 | myo2-E1 myp2-Δ1∷his7+ spn1-Δ2 ade6 his7-366 leu1-32 ura4-D18 | This studym |
| JW2613 | h+/h− scw1-ng124/scw1+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 | JW2271 × JW81 |
| JW2614 | h+/h− scw1-ng124/scw1Δ∷ura4+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 | DM1274 × JW2271 |
Strain . | Genotypea . | Source/reference . |
|---|---|---|
| 972 | h− wild type | Leupold (1970) |
| DM1274 | h− scw1Δ∷ura4+ ade6-M210 leu1-32 ura4-D18 | Jin and McCollum (2003) |
| FY435 | h+ ade6-M210 his7-366 leu1-32 ura4-D18 | S. Forsburg |
| FY11065 | h− cds1-2HA-6His∷ura4+ leu1 ura4 | Yeast Genetic Resource Center Japan |
| KS3320 | h+ swi5Δ∷kanMX6 | Anders et al. (2008) |
| KS3250 | h− rec12Δ∷kanMX6 ade6-M210 his7-366 leu1-32 lys1-37 | Anders et al. (2008) |
| MBY580 | h− bgs1-191 ade6-M210 leu1-32 lys1-131 ura4-D18 | Liu et al. (1999)b |
| TP5 | h− myp2Δ∷his7+ ade6-M210 his7-366 leu1-32 ura4-D18 | Bezanilla et al. (1997) |
| TP90 | h+ myo2-E1 myp2-Δ1∷his7+ade6-M216 his7-366 leu1-32 ura4-D18 | M. Bezanilla and T. Pollard |
| YDM74 | h− myo2-E1 ade6 his3-D1 leu1-32 ura4-D18 | D. McCollumc |
| JW21 | h+ cdc4-8 | P. Nurse; Nurse et al. (1976); McCollum et al. (1995) |
| JW81 | h− ade6-M210 leu1-32 ura4-D18 | Lab collection |
| JW182 | h− spn4-Δ2 | This studyd |
| JW183 | h− spn4+-GFP | This studyd |
| JW215 | h+ his3-27 leu1-32 ura4-D18 | Lab collection |
| JW249 | h− spn1-Δ2 | This studyd |
| JW251 | h− spn1+-GFP | This studyd |
| JW267 | h− 81nmt1-spn1+ 81nmt1-spn4+ leu1-32 ura4-D18 | See text. |
| JW289 | h+ spn1-Δ2 leu1-32 ura4-D18 | JW215 × JW249 |
| JW290 | h− spn1-Δ2 his3-27 ura4-D18 | JW215 × JW249 |
| JW295 | h+ spn4-Δ2 leu1-32 ura4-D18 | JW215 × JW182 |
| JW306 | h+ spn1+-GFP his3-27 leu1-32 ura4-D18 | JW215 × JW251 |
| JW314 | spn4-Δ2 mid2Δ∷ura4+ leu1-32 ura4-D18 | JW295 × JW430 |
| JW315 | scw1-ng124 mid2Δ∷ura4+ leu1-32 ura4-D18 | ng124e × JW430 |
| JW318 | spn1+-GFP mid2Δ∷ura4+ leu1-32 ura4-D18 | JW306 × JW430 |
| JW320 | scw1-ng124 spn4+-GFP leu1-32 ura4-D18 | JW331 × JW183 |
| JW321-1 | scw1-ng124 spn1-Δ2 ura4-D18 | JW331 × JW290 |
| JW326 | h− mid2+-GFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW330 | h− scw1-ng124 ade6-M210 leu1-32 ura4-D18 | This studyf |
| JW331 | h+ scw1-ng124 ade6-M210 leu1-32 ura4-D18 | This studyf |
| JW332 | mid2+-GFP spn4-Δ2 leu1-32 ura4-D18 | JW326 × JW295 |
| JW345 | cdc4-s16 myp2Δ∷his7+ ade6-M210 his7-366 leu1-32 ura4-D18 | JW400 × TP5 |
| JW374 | art1-s34 spn1-Δ2 leu1-32 ura4-D18 | JW403 × JW289 |
| JW380 | h− myp2Δ∷his7+ spn1-Δ2 ade6 his7-366 leu1-32 ura4-D18 | This studyg |
| JW396 | h− s26 leu1-32 ura4-D18 | This studyh |
| JW397 | h− s63 his3-27 leu1-32 ura4-D18 | This studyh |
| JW398 | h+ s28 leu1-32 ura4-D18 | This studyh |
| JW399 | h− cdc4-s16 leu1-32 ura4-D18 | This studye |
| JW400 | h+ cdc4-s16 ade6-M210 his7-366 leu1-32 ura4-D18 | JW399 × FY435 |
| JW401 | cdc4-s16 spn4-Δ2 leu1-32 ura4-D18 | JW399 × JW295 |
| JW402 | h− art1-s34 81nmt1-spn1+ 81nmt1-spn4+ leu1-32 ura4-D18 | This studyi |
| JW403 | h− art1-s34 leu1-32 ura4-D18 | This studye |
| JW404 | h+ art1-s34 leu1-32 ura4-D18 | This studye |
| JW405 | h− rgf3-s44 81nmt1-spn1+ 81nmt1-spn4+ leu1-32 ura4-D18 | This studyi |
| JW406 | h− rgf3-s44 his3-27 leu1-32 ura4-D18 | This studye |
| JW407 | h+ rgf3-s44 his3-27 leu1-32 ura4-D18 | This studye |
| JW409 | h− rgf1-GFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW412 | h− rgf3-GFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW423 | h− rgf1-Δ1 ade6 leu1-32 ura4-D18 | This studyj |
| JW430 | h− mid2Δ∷ura4+ade6 leu1-32 ura4-D18 | D. Brunner and P. Nurse |
| JW729 | h+ ade6-M210 leu1-32 ura4-D18 | Lab collection |
| JW903 | h+ 3nmt1-rgf2 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1078 | h− 3nmt1-rgf1 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1081 | h− 3nmt1-rgf3 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1084 | h− sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1100 | h+ spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1105 | h+ rgf3-mEGFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1113 | h− spn1-mEGFP sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | JW1100 × JW1084 |
| JW1122 | 3nmt1-rgf3 spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | JW1081 × JW1100 |
| JW1123 | 3nmt1-rgf1 spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | JW1078 × JW1100 |
| JW1124 | rgf1-GFP sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | This studyk |
| JW1125 | rgf3-s44 spn1-mEGFP leu1-32 ura4-D18 | JW406 × JW1100 |
| JW1126 | art1-s34 spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | JW403 × JW1100 |
| JW1128 | rgf3-GFP spn1-Δ2 ade6-M210 leu1-32 ura4-D18 | JW412 × JW289 |
| JW1131 | rgf3-mEGFP sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | JW1105 × JW1084 |
| JW1139 | rgf1-GFP spn1-Δ2 ade6-M210 leu1-32 ura4-D18 | JW409 × JW289 |
| JW1203 | mid2-GFP scw1-ng124 ade6-M210 leu1-32 ura4-D18 | JW326 × JW331 |
| JW2068 | h+ sec15Δ∷kanMX4 ade6 leu1-32 ura4-D18 | Lab collection |
| JW2155 | h− scw1-ng124 ade6-M210 leu1-32 ura4-D18 | This studyl |
| JW2271 | h+ scw1-ng124 ade6-M216 leu1-32 ura4-D18 | This studyl |
| JW2286 | scw1Δ∷ura4+ spn1-Δ2 ade6-M210 leu1-32 ura4-D18 | DM1274 × JW289 |
| JW2499 | cdc4-s16 scw1-ng124 ade6 leu1-32 ura4-D18 | JW399 × JW2271 |
| JW2503 | myo2-E1 scw1-ng124 ade6 leu1-32 ura4-D18 | YDM74 × JW2271 |
| JW2560 | myo2-E1 myp2-Δ1∷his7+ spn1-Δ2 ade6 his7-366 leu1-32 ura4-D18 | This studym |
| JW2613 | h+/h− scw1-ng124/scw1+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 | JW2271 × JW81 |
| JW2614 | h+/h− scw1-ng124/scw1Δ∷ura4+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 | DM1274 × JW2271 |
All deletions and taggings constructed in this study by the PCR method used the kanMX6 marker, which is not indicated in the genotypes. The mating types of some strains were not determined. GFP(S65T) is indicated simply as GFP; mEGFP (see materials and methods) is indicated as such.
bgs1-191 was originally known as drc1-191 (Liu et al. 1999).
myo2-E1 was originally referred to as rng5-E1 (Balasubramanian et al. 1998; Eng et al. 1998).
Strain 972 was transformed with the appropriate constructs (see text).
Segregant from the third backcross of the original mutant isolate to strains JW81 and JW215.
Segregant from the fourth backcross of the original mutant isolate to strains JW81 and JW215.
Derived by several crosses from strains TP5 and JW249.
Segregant from the first backcross of the original mutant isolate to strain JW215.
The original mutant isolate.
Segregant from a heterozygous mutant obtained after PCR-mediated transformation (see text) of diploid strain JW9 (Wu et al. 2001).
Derived by crossing JW409 to a segregant from the cross of JW1084 to JW729 (Wu et al. 2003).
Segregant from the 10th backcross of the original mutant isolate to strains JW81 and JW729.
Derived by several crosses from strains TP5, YDM74, and JW249.
S. pombe strains used in this study
Strain . | Genotypea . | Source/reference . |
|---|---|---|
| 972 | h− wild type | Leupold (1970) |
| DM1274 | h− scw1Δ∷ura4+ ade6-M210 leu1-32 ura4-D18 | Jin and McCollum (2003) |
| FY435 | h+ ade6-M210 his7-366 leu1-32 ura4-D18 | S. Forsburg |
| FY11065 | h− cds1-2HA-6His∷ura4+ leu1 ura4 | Yeast Genetic Resource Center Japan |
| KS3320 | h+ swi5Δ∷kanMX6 | Anders et al. (2008) |
| KS3250 | h− rec12Δ∷kanMX6 ade6-M210 his7-366 leu1-32 lys1-37 | Anders et al. (2008) |
| MBY580 | h− bgs1-191 ade6-M210 leu1-32 lys1-131 ura4-D18 | Liu et al. (1999)b |
| TP5 | h− myp2Δ∷his7+ ade6-M210 his7-366 leu1-32 ura4-D18 | Bezanilla et al. (1997) |
| TP90 | h+ myo2-E1 myp2-Δ1∷his7+ade6-M216 his7-366 leu1-32 ura4-D18 | M. Bezanilla and T. Pollard |
| YDM74 | h− myo2-E1 ade6 his3-D1 leu1-32 ura4-D18 | D. McCollumc |
| JW21 | h+ cdc4-8 | P. Nurse; Nurse et al. (1976); McCollum et al. (1995) |
| JW81 | h− ade6-M210 leu1-32 ura4-D18 | Lab collection |
| JW182 | h− spn4-Δ2 | This studyd |
| JW183 | h− spn4+-GFP | This studyd |
| JW215 | h+ his3-27 leu1-32 ura4-D18 | Lab collection |
| JW249 | h− spn1-Δ2 | This studyd |
| JW251 | h− spn1+-GFP | This studyd |
| JW267 | h− 81nmt1-spn1+ 81nmt1-spn4+ leu1-32 ura4-D18 | See text. |
| JW289 | h+ spn1-Δ2 leu1-32 ura4-D18 | JW215 × JW249 |
| JW290 | h− spn1-Δ2 his3-27 ura4-D18 | JW215 × JW249 |
| JW295 | h+ spn4-Δ2 leu1-32 ura4-D18 | JW215 × JW182 |
| JW306 | h+ spn1+-GFP his3-27 leu1-32 ura4-D18 | JW215 × JW251 |
| JW314 | spn4-Δ2 mid2Δ∷ura4+ leu1-32 ura4-D18 | JW295 × JW430 |
| JW315 | scw1-ng124 mid2Δ∷ura4+ leu1-32 ura4-D18 | ng124e × JW430 |
| JW318 | spn1+-GFP mid2Δ∷ura4+ leu1-32 ura4-D18 | JW306 × JW430 |
| JW320 | scw1-ng124 spn4+-GFP leu1-32 ura4-D18 | JW331 × JW183 |
| JW321-1 | scw1-ng124 spn1-Δ2 ura4-D18 | JW331 × JW290 |
| JW326 | h− mid2+-GFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW330 | h− scw1-ng124 ade6-M210 leu1-32 ura4-D18 | This studyf |
| JW331 | h+ scw1-ng124 ade6-M210 leu1-32 ura4-D18 | This studyf |
| JW332 | mid2+-GFP spn4-Δ2 leu1-32 ura4-D18 | JW326 × JW295 |
| JW345 | cdc4-s16 myp2Δ∷his7+ ade6-M210 his7-366 leu1-32 ura4-D18 | JW400 × TP5 |
| JW374 | art1-s34 spn1-Δ2 leu1-32 ura4-D18 | JW403 × JW289 |
| JW380 | h− myp2Δ∷his7+ spn1-Δ2 ade6 his7-366 leu1-32 ura4-D18 | This studyg |
| JW396 | h− s26 leu1-32 ura4-D18 | This studyh |
| JW397 | h− s63 his3-27 leu1-32 ura4-D18 | This studyh |
| JW398 | h+ s28 leu1-32 ura4-D18 | This studyh |
| JW399 | h− cdc4-s16 leu1-32 ura4-D18 | This studye |
| JW400 | h+ cdc4-s16 ade6-M210 his7-366 leu1-32 ura4-D18 | JW399 × FY435 |
| JW401 | cdc4-s16 spn4-Δ2 leu1-32 ura4-D18 | JW399 × JW295 |
| JW402 | h− art1-s34 81nmt1-spn1+ 81nmt1-spn4+ leu1-32 ura4-D18 | This studyi |
| JW403 | h− art1-s34 leu1-32 ura4-D18 | This studye |
| JW404 | h+ art1-s34 leu1-32 ura4-D18 | This studye |
| JW405 | h− rgf3-s44 81nmt1-spn1+ 81nmt1-spn4+ leu1-32 ura4-D18 | This studyi |
| JW406 | h− rgf3-s44 his3-27 leu1-32 ura4-D18 | This studye |
| JW407 | h+ rgf3-s44 his3-27 leu1-32 ura4-D18 | This studye |
| JW409 | h− rgf1-GFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW412 | h− rgf3-GFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW423 | h− rgf1-Δ1 ade6 leu1-32 ura4-D18 | This studyj |
| JW430 | h− mid2Δ∷ura4+ade6 leu1-32 ura4-D18 | D. Brunner and P. Nurse |
| JW729 | h+ ade6-M210 leu1-32 ura4-D18 | Lab collection |
| JW903 | h+ 3nmt1-rgf2 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1078 | h− 3nmt1-rgf1 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1081 | h− 3nmt1-rgf3 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1084 | h− sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1100 | h+ spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1105 | h+ rgf3-mEGFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1113 | h− spn1-mEGFP sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | JW1100 × JW1084 |
| JW1122 | 3nmt1-rgf3 spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | JW1081 × JW1100 |
| JW1123 | 3nmt1-rgf1 spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | JW1078 × JW1100 |
| JW1124 | rgf1-GFP sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | This studyk |
| JW1125 | rgf3-s44 spn1-mEGFP leu1-32 ura4-D18 | JW406 × JW1100 |
| JW1126 | art1-s34 spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | JW403 × JW1100 |
| JW1128 | rgf3-GFP spn1-Δ2 ade6-M210 leu1-32 ura4-D18 | JW412 × JW289 |
| JW1131 | rgf3-mEGFP sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | JW1105 × JW1084 |
| JW1139 | rgf1-GFP spn1-Δ2 ade6-M210 leu1-32 ura4-D18 | JW409 × JW289 |
| JW1203 | mid2-GFP scw1-ng124 ade6-M210 leu1-32 ura4-D18 | JW326 × JW331 |
| JW2068 | h+ sec15Δ∷kanMX4 ade6 leu1-32 ura4-D18 | Lab collection |
| JW2155 | h− scw1-ng124 ade6-M210 leu1-32 ura4-D18 | This studyl |
| JW2271 | h+ scw1-ng124 ade6-M216 leu1-32 ura4-D18 | This studyl |
| JW2286 | scw1Δ∷ura4+ spn1-Δ2 ade6-M210 leu1-32 ura4-D18 | DM1274 × JW289 |
| JW2499 | cdc4-s16 scw1-ng124 ade6 leu1-32 ura4-D18 | JW399 × JW2271 |
| JW2503 | myo2-E1 scw1-ng124 ade6 leu1-32 ura4-D18 | YDM74 × JW2271 |
| JW2560 | myo2-E1 myp2-Δ1∷his7+ spn1-Δ2 ade6 his7-366 leu1-32 ura4-D18 | This studym |
| JW2613 | h+/h− scw1-ng124/scw1+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 | JW2271 × JW81 |
| JW2614 | h+/h− scw1-ng124/scw1Δ∷ura4+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 | DM1274 × JW2271 |
Strain . | Genotypea . | Source/reference . |
|---|---|---|
| 972 | h− wild type | Leupold (1970) |
| DM1274 | h− scw1Δ∷ura4+ ade6-M210 leu1-32 ura4-D18 | Jin and McCollum (2003) |
| FY435 | h+ ade6-M210 his7-366 leu1-32 ura4-D18 | S. Forsburg |
| FY11065 | h− cds1-2HA-6His∷ura4+ leu1 ura4 | Yeast Genetic Resource Center Japan |
| KS3320 | h+ swi5Δ∷kanMX6 | Anders et al. (2008) |
| KS3250 | h− rec12Δ∷kanMX6 ade6-M210 his7-366 leu1-32 lys1-37 | Anders et al. (2008) |
| MBY580 | h− bgs1-191 ade6-M210 leu1-32 lys1-131 ura4-D18 | Liu et al. (1999)b |
| TP5 | h− myp2Δ∷his7+ ade6-M210 his7-366 leu1-32 ura4-D18 | Bezanilla et al. (1997) |
| TP90 | h+ myo2-E1 myp2-Δ1∷his7+ade6-M216 his7-366 leu1-32 ura4-D18 | M. Bezanilla and T. Pollard |
| YDM74 | h− myo2-E1 ade6 his3-D1 leu1-32 ura4-D18 | D. McCollumc |
| JW21 | h+ cdc4-8 | P. Nurse; Nurse et al. (1976); McCollum et al. (1995) |
| JW81 | h− ade6-M210 leu1-32 ura4-D18 | Lab collection |
| JW182 | h− spn4-Δ2 | This studyd |
| JW183 | h− spn4+-GFP | This studyd |
| JW215 | h+ his3-27 leu1-32 ura4-D18 | Lab collection |
| JW249 | h− spn1-Δ2 | This studyd |
| JW251 | h− spn1+-GFP | This studyd |
| JW267 | h− 81nmt1-spn1+ 81nmt1-spn4+ leu1-32 ura4-D18 | See text. |
| JW289 | h+ spn1-Δ2 leu1-32 ura4-D18 | JW215 × JW249 |
| JW290 | h− spn1-Δ2 his3-27 ura4-D18 | JW215 × JW249 |
| JW295 | h+ spn4-Δ2 leu1-32 ura4-D18 | JW215 × JW182 |
| JW306 | h+ spn1+-GFP his3-27 leu1-32 ura4-D18 | JW215 × JW251 |
| JW314 | spn4-Δ2 mid2Δ∷ura4+ leu1-32 ura4-D18 | JW295 × JW430 |
| JW315 | scw1-ng124 mid2Δ∷ura4+ leu1-32 ura4-D18 | ng124e × JW430 |
| JW318 | spn1+-GFP mid2Δ∷ura4+ leu1-32 ura4-D18 | JW306 × JW430 |
| JW320 | scw1-ng124 spn4+-GFP leu1-32 ura4-D18 | JW331 × JW183 |
| JW321-1 | scw1-ng124 spn1-Δ2 ura4-D18 | JW331 × JW290 |
| JW326 | h− mid2+-GFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW330 | h− scw1-ng124 ade6-M210 leu1-32 ura4-D18 | This studyf |
| JW331 | h+ scw1-ng124 ade6-M210 leu1-32 ura4-D18 | This studyf |
| JW332 | mid2+-GFP spn4-Δ2 leu1-32 ura4-D18 | JW326 × JW295 |
| JW345 | cdc4-s16 myp2Δ∷his7+ ade6-M210 his7-366 leu1-32 ura4-D18 | JW400 × TP5 |
| JW374 | art1-s34 spn1-Δ2 leu1-32 ura4-D18 | JW403 × JW289 |
| JW380 | h− myp2Δ∷his7+ spn1-Δ2 ade6 his7-366 leu1-32 ura4-D18 | This studyg |
| JW396 | h− s26 leu1-32 ura4-D18 | This studyh |
| JW397 | h− s63 his3-27 leu1-32 ura4-D18 | This studyh |
| JW398 | h+ s28 leu1-32 ura4-D18 | This studyh |
| JW399 | h− cdc4-s16 leu1-32 ura4-D18 | This studye |
| JW400 | h+ cdc4-s16 ade6-M210 his7-366 leu1-32 ura4-D18 | JW399 × FY435 |
| JW401 | cdc4-s16 spn4-Δ2 leu1-32 ura4-D18 | JW399 × JW295 |
| JW402 | h− art1-s34 81nmt1-spn1+ 81nmt1-spn4+ leu1-32 ura4-D18 | This studyi |
| JW403 | h− art1-s34 leu1-32 ura4-D18 | This studye |
| JW404 | h+ art1-s34 leu1-32 ura4-D18 | This studye |
| JW405 | h− rgf3-s44 81nmt1-spn1+ 81nmt1-spn4+ leu1-32 ura4-D18 | This studyi |
| JW406 | h− rgf3-s44 his3-27 leu1-32 ura4-D18 | This studye |
| JW407 | h+ rgf3-s44 his3-27 leu1-32 ura4-D18 | This studye |
| JW409 | h− rgf1-GFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW412 | h− rgf3-GFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW423 | h− rgf1-Δ1 ade6 leu1-32 ura4-D18 | This studyj |
| JW430 | h− mid2Δ∷ura4+ade6 leu1-32 ura4-D18 | D. Brunner and P. Nurse |
| JW729 | h+ ade6-M210 leu1-32 ura4-D18 | Lab collection |
| JW903 | h+ 3nmt1-rgf2 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1078 | h− 3nmt1-rgf1 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1081 | h− 3nmt1-rgf3 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1084 | h− sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1100 | h+ spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1105 | h+ rgf3-mEGFP ade6-M210 leu1-32 ura4-D18 | See text. |
| JW1113 | h− spn1-mEGFP sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | JW1100 × JW1084 |
| JW1122 | 3nmt1-rgf3 spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | JW1081 × JW1100 |
| JW1123 | 3nmt1-rgf1 spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | JW1078 × JW1100 |
| JW1124 | rgf1-GFP sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | This studyk |
| JW1125 | rgf3-s44 spn1-mEGFP leu1-32 ura4-D18 | JW406 × JW1100 |
| JW1126 | art1-s34 spn1-mEGFP ade6-M210 leu1-32 ura4-D18 | JW403 × JW1100 |
| JW1128 | rgf3-GFP spn1-Δ2 ade6-M210 leu1-32 ura4-D18 | JW412 × JW289 |
| JW1131 | rgf3-mEGFP sad1-mRFP1 ade6-M210 leu1-32 ura4-D18 | JW1105 × JW1084 |
| JW1139 | rgf1-GFP spn1-Δ2 ade6-M210 leu1-32 ura4-D18 | JW409 × JW289 |
| JW1203 | mid2-GFP scw1-ng124 ade6-M210 leu1-32 ura4-D18 | JW326 × JW331 |
| JW2068 | h+ sec15Δ∷kanMX4 ade6 leu1-32 ura4-D18 | Lab collection |
| JW2155 | h− scw1-ng124 ade6-M210 leu1-32 ura4-D18 | This studyl |
| JW2271 | h+ scw1-ng124 ade6-M216 leu1-32 ura4-D18 | This studyl |
| JW2286 | scw1Δ∷ura4+ spn1-Δ2 ade6-M210 leu1-32 ura4-D18 | DM1274 × JW289 |
| JW2499 | cdc4-s16 scw1-ng124 ade6 leu1-32 ura4-D18 | JW399 × JW2271 |
| JW2503 | myo2-E1 scw1-ng124 ade6 leu1-32 ura4-D18 | YDM74 × JW2271 |
| JW2560 | myo2-E1 myp2-Δ1∷his7+ spn1-Δ2 ade6 his7-366 leu1-32 ura4-D18 | This studym |
| JW2613 | h+/h− scw1-ng124/scw1+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 | JW2271 × JW81 |
| JW2614 | h+/h− scw1-ng124/scw1Δ∷ura4+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 | DM1274 × JW2271 |
All deletions and taggings constructed in this study by the PCR method used the kanMX6 marker, which is not indicated in the genotypes. The mating types of some strains were not determined. GFP(S65T) is indicated simply as GFP; mEGFP (see materials and methods) is indicated as such.
bgs1-191 was originally known as drc1-191 (Liu et al. 1999).
myo2-E1 was originally referred to as rng5-E1 (Balasubramanian et al. 1998; Eng et al. 1998).
Strain 972 was transformed with the appropriate constructs (see text).
Segregant from the third backcross of the original mutant isolate to strains JW81 and JW215.
Segregant from the fourth backcross of the original mutant isolate to strains JW81 and JW215.
Derived by several crosses from strains TP5 and JW249.
Segregant from the first backcross of the original mutant isolate to strain JW215.
The original mutant isolate.
Segregant from a heterozygous mutant obtained after PCR-mediated transformation (see text) of diploid strain JW9 (Wu et al. 2001).
Derived by crossing JW409 to a segregant from the cross of JW1084 to JW729 (Wu et al. 2003).
Segregant from the 10th backcross of the original mutant isolate to strains JW81 and JW729.
Derived by several crosses from strains TP5, YDM74, and JW249.
Standard rich (YE5S) and minimal (EMM) media and genetic methods were used (Moreno et al. 1991); EMM5S medium was EMM containing also adenine, histidine, leucine, lysine, and uracil. To repress expression from the nmt1 promoter, 5 μg/ml thiamine was added to the medium. Some solid media contained 2.5 μg/ml phloxin B, which accumulates in dead cells to give a red colony color (Moreno et al. 1991). Standard molecular biology methods were used except where noted. Oligonucleotide primers were purchased from Integrated DNA Technologies.
Mutagenesis and screening for synthetic-effect mutations:
To regulate the expression levels of spn1+ and spn4+, their promoters were replaced with the attenuated 81nmt1 promoter in wild-type strain 972 by the PCR method (Bähler et al. 1998) using the kanMX6 marker, forward primers corresponding to nucleotides −111 to −42 (spn1) or −134 to −55 (spn4) relative to the start codons, and reverse primers based on the N-terminal codons. In both cases, the upstream ORFs are far enough away that these manipulations are unlikely to affect their expression. Checking by PCR identified transformants (JW254 and JW188) that had sustained the desired integrations, and G418 resistance segregated 2:2 in crosses of these strains to JW215. Appropriate segregants were then crossed to obtain strain JW267 (81nmt1-spn1+ 81nmt1-spn4+).
To screen for synthetic-effect mutations, strain JW267 was grown in EMM5S to ∼107 cells/ml, mutagenized with 300 μg/ml nitrosoguanidine (Sigma-Aldrich) at 30° for 45 min (∼20% survival) as described by Moreno et al. (1991), and plated onto EMM5S (∼500 viable cells/plate) at 30°. The resulting colonies were replica-plated onto EMM5S + phloxin (inducing conditions) and EMM5S + thiamine + phloxin (repressing conditions) at 30°. Colonies that were significantly redder or grew significantly less well on the thiamine plates were restreaked under inducing and repressing conditions. Possible mutants with modest growth defects were also screened microscopically to identify those with strong morphological phenotypes. Before detailed characterization, each mutant was backcrossed three or more times to wild-type strains JW81, JW215, and/or JW729 to obtain strains that carried the new mutations without 81nmt1-spn1 or 81nmt1-spn4.
Identification of the genes defined by synthetic-effect mutations:
To clone the gene defined by mutation s16, spn4Δ s16 strain JW401 was transformed with the pWH5 and pURSP2 libraries, grown at 36° on EMM5S-Leu or EMM5S-Ura, and replica-plated onto phloxin-containing plates at 23°. Three transformants with reduced phloxin uptake were found. After recovery in Escherichia coli, all three plasmids could rescue the mutant phenotypes of strains JW401 and JW399 (s16) to spn4Δ-like and wild-type phenotypes, respectively, at 23°. Sequencing revealed that the plasmids all contained the complete cdc4 coding sequence and the C-terminal ∼190 codons of the adjacent, 590-codon paa1. To ask if s16 was a mutation in cdc4, a genomic DNA fragment that extended from nucleotide −197 upstream to nucleotide 173 downstream of the cdc4 coding region was amplified from strain JW399 using the Expand High Fidelity PCR system (Roche). The PCR product was purified using Qiagen columns and sequenced.
To clone the genes defined by mutations s34 and s44, strains JW402 (s34 81nmt1-spn1+ 81nmt1-spn4+) and JW405 (s44 81nmt1-spn1+ 81nmt1-spn4+) were grown in EMM5S medium, transformed with the pURSP1 and pURSP2 libraries, and plated on EMM5S-Ura + thiamine at 30°. Plasmids from large colonies were recovered into E. coli, and those that could at least partially rescue the growth defects and cell-lysis phenotypes of the mutant strains upon retransformation were classified by restriction digestion and partial sequencing (see results). To identify the site of the s34 mutation, genomic DNA fragments from strain JW404 were amplified using the Expand or iProof (Bio-Rad) high-fidelity PCR system, purified on Qiagen columns, and sequenced. Collectively, the fragments covered 25,089 bp around bgs1 on chromosome II (nucleotides 2,341,116–2,366,204). The same approach was used to identify the s44 mutation in strain JW407. The region sequenced covered 10,321 bp (nucleotides 1,239,244–1,249,564) on chromosome III, which includes the entire ORFs of the adjacent and divergently transcribed rgf1 and rgf3, the 2050-bp region between them, and 141 and 251 bp of their respective 3′ UTRs. In both cases, the mutant sites were confirmed by sequencing several fragments from independent PCR reactions.
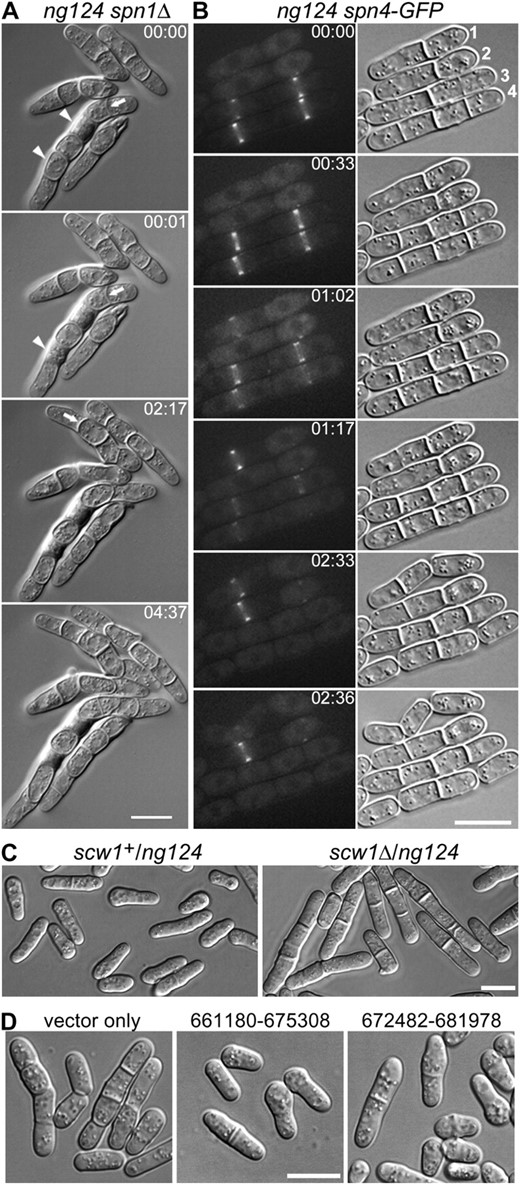
To identify the gene defined by mutation ng124, we used positional cloning. First, we used strains containing a rec12Δ mutation [which reduces homologous recombination by ∼1000-fold when homozygous (Anders et al. 2008)] and markers on each chromosome. This revealed cosegregation of ng124 with ade6+, indicating that ng124 is on chromosome III. Next, we used strains carrying swi5Δ [which reduces recombination by ∼10-fold when homozygous (Schmidt 1993)], revealing linkage of ng124 to ura4 (∼15 cM in the swi5Δ background). Crosses using swi5+ strains then showed that ng124 was ∼7 cM from sec15 and ∼12 cM from cds1, suggesting that ng124 lies between these two genes (which are ∼20 cM apart). One gene in this region is scw1, mutations in which have been shown also to produce a multi-septum phenotype (Karagiannis et al. 2002; Jin and McCollum 2003). Several lines of evidence suggested that ng124 is indeed a mutation in scw1 (see results), and this was confirmed by sequencing genomic DNA (chromosome III nucleotides 674,476–675,161) from an extensively backcrossed ng124 strain (JW2155), which revealed a mutation in scw1 codon 276 (see results).
Gene deletion and tagging:
Genes were deleted (from start codon to stop codon) or tagged at the C termini of their chromosomal loci by the PCR method (Bähler et al. 1998; Wu et al. 2001) using the kanMX6 marker. The tags encoded GFP (green fluorescent protein with the S65T mutation), mEGFP [GFP with the F64L and S65T mutations to enhance fluorescence and the A206K mutation to minimize dimerization (Yang et al. 1996; Zacharias et al. 2002; Wu et al. 2003)], or mRFP1 [monomeric red fluorescent protein (Campbell et al. 2002; Huh et al. 2003)]. The gene-specific sequences of the forward primers ended immediately upstream of the stop codons, and those of the reverse primers ended short distances downstream of the stop codons (1 bp for rgf1, 10 bp for rgf2, 8 bp for rgf3, 28 bp for spn1, 44 bp for spn4, 5 bp for mid2, and 1 bp for sad1). All of the tagged proteins appeared to be functional as judged by normal colony growth and normal cell morphologies in the tagged strains (Wu et al. 2003; data not shown). The PCR method was also used to place genes under the control of the strong (3nmt1) nmt1 promoter (Basi et al. 1993; Forsburg 1993); the gene-specific sequences of the forward primers ended short distances upstream of the start codons (5 bp for rgf1 and 6 bp for rgf2 and rgf3) and those of the reverse primers ended immediately downstream of the start codons. Transformants were recovered on YE5S + thiamine to repress the nmt1 promoter.
Morphological observations:
In most cases, cells were observed by fluorescence or differential-interference-contrast (DIC) microscopy using either a Nikon Eclipse 600 FN microscope with a Plan-Apo 100X/1.4 NA objective and a Hamamatsu ORCA-2 cooled CCD camera or an Olympus IX-71 inverted microscope with a Plan-Apo 60X/1.4 NA objective and an ORCA-ER cooled CCD camera. The three-dimensional images in Figure 6 were obtained using an UltraView RS spinning-disk confocal microscope (Perkin Elmer Life Sciences) and a 488-nm argon ion laser. Septa and DNA were stained using Calcofluor and bisbenzamide as described by Wu et al. (2001). For time-lapse experiments, cells were pre-grown in the appropriate medium to ∼4 × 106 cells/ml, mounted in growth chambers prepared as described by Maddox et al. (2000) and Wu et al. (2003), and observed at 24° or 25°. Images were collected at 30-sec to 2-min intervals using exposures of 0.065–0.3 sec (DIC) or 0.1–3 sec (GFP) and analyzed using MetaMorph (Molecular Devices) or Image J (http://rsb.info.nih.gov/ij/) software.
RESULTS
Identification of synthetic-effect mutations:
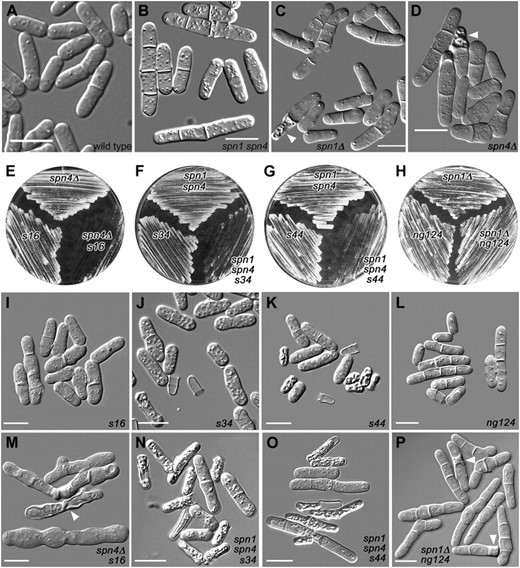
To identify proteins or pathways that might be redundant with the septins, we sought mutations that were lethal (or nearly so) when the septins were absent. We used a strain in which both spn1+ and spn4+ are under control of the weak, thiamine-repressible 81nmt1 promoter. Under inducing conditions, these cells resembled wild type, but under repressing conditions, they resembled spn1 and spn4 deletion mutants: the growth rate was close to that of wild type, but many more cells than normal had visible septa (and sometimes several septa), indicating a delay in septum completion and/or cell separation (Figure 1, A–D). We mutagenized with nitrosoguanidine and screened ∼150,000 colonies, obtaining seven mutants that died or grew poorly under repressing conditions (see materials and methods). Strains containing only the new mutations were isolated by backcrossing to wild type, and all had defects detectable by phloxin staining, by microscopic examination, or both (see below). Each mutant segregated 2:2 for this phenotype, indicating that a single mutation was involved. Data presented below show that mutation s16 is in cdc4. Linkage analyses (Table 2, lines 1–3 and 5–13) indicated that mutations s34, s26, and s63 are tightly linked to each other and therefore likely to define a second gene; that mutations s44 and s28 are tightly linked to each other, but not to s34, s26, and s63, and therefore likely to define a third gene; and that mutation ng124 defines a fourth gene that is not linked to any of the other three.
Mutations showing synthetic effects with septin mutations. Cells were observed directly in liquid culture medium by DIC microscopy (A–D and I–P) or streaked on plates to observe overall growth rates (E–H). (A–D) Wild-type strain JW81 and the septin-mutant strains JW267, JW289, and JW182. (I–L) Strains carrying the new mutations in otherwise wild-type backgrounds (JW399, JW403, JW406, and JW330). (M–P) The double or triple mutants (JW401, JW402, JW405, and JW321-1). Plates show the same strains. spn1 spn4 designates 81nmt1-spn1+ 81nmt1-spn4+. (B, J, K, N, and O) Cells were grown in EMM5S without thiamine and then shifted to EMM5S + thiamine at 30° for 24 hr to repress the nmt1 promoters. (A, C, L, and P) Cells were grown in YE5S at 30°. (D, I, and M) Cells were grown overnight in EMM5S at 32° and then shifted to 23° for 8 hr. (E) Cells were grown on YE5S at 23° for 5 days. (F and G) Cells were grown on EMM5S + thiamine at 36° for 3 days. (H) Cells were grown on YE5S at 30° for 2 days. Arrowheads in C, D, M, and P indicate cells that appear to have lysed; such cells are numerous in J, K, N, and O. Bars, 10 μm.
Linkage analysis of mutations identified in the synthetic-effect screen
. | Tetrad type . | |||
|---|---|---|---|---|
| Line . | Cross . | PD . | NPD . | T . |
| 1 | s34 × s26 | 41 | 0 | 0 |
| 2 | s34 × s63 | 43 | 0 | 0 |
| 3 | s44 × s28 | 37 | 0 | 0 |
| 4 | s28 × rgf1-Δ1a | 20 | 0 | 0 |
| 5 | s34 × s28 | 1 | 5 | 4 |
| 6 | s34 × s44 | 1 | 0 | 6 |
| 7 | s26 × s28 | 2 | 0 | 7 |
| 8 | s63 × s28 | 1 | 1 | 8 |
| 9 | ng124 × s16 | 2 | 0 | 7 |
| 10 | ng124 × s26 | 2 | 1 | 7 |
| 11 | ng124 × s34 | 0 | 0 | 3 |
| 12 | ng124 × s63 | 1 | 4 | 8 |
| 13 | ng124 × s44 | 2 | 0 | 5 |
. | Tetrad type . | |||
|---|---|---|---|---|
| Line . | Cross . | PD . | NPD . | T . |
| 1 | s34 × s26 | 41 | 0 | 0 |
| 2 | s34 × s63 | 43 | 0 | 0 |
| 3 | s44 × s28 | 37 | 0 | 0 |
| 4 | s28 × rgf1-Δ1a | 20 | 0 | 0 |
| 5 | s34 × s28 | 1 | 5 | 4 |
| 6 | s34 × s44 | 1 | 0 | 6 |
| 7 | s26 × s28 | 2 | 0 | 7 |
| 8 | s63 × s28 | 1 | 1 | 8 |
| 9 | ng124 × s16 | 2 | 0 | 7 |
| 10 | ng124 × s26 | 2 | 1 | 7 |
| 11 | ng124 × s34 | 0 | 0 | 3 |
| 12 | ng124 × s63 | 1 | 4 | 8 |
| 13 | ng124 × s44 | 2 | 0 | 5 |
Appropriate single-mutant strains (Table 1) were crossed, and tetrads were dissected and incubated at 30° or 32°. For crosses involving s44, spores were germinated on YE5S + 1 m sorbitol to reduce cell lysis. Mutant and wild-type segregants were distinguished by growing on YE5S medium containing phloxin B or (for ng124) by checking the morphological phenotypes microscopically.
Strain JW423 (Table 1).
Linkage analysis of mutations identified in the synthetic-effect screen
. | Tetrad type . | |||
|---|---|---|---|---|
| Line . | Cross . | PD . | NPD . | T . |
| 1 | s34 × s26 | 41 | 0 | 0 |
| 2 | s34 × s63 | 43 | 0 | 0 |
| 3 | s44 × s28 | 37 | 0 | 0 |
| 4 | s28 × rgf1-Δ1a | 20 | 0 | 0 |
| 5 | s34 × s28 | 1 | 5 | 4 |
| 6 | s34 × s44 | 1 | 0 | 6 |
| 7 | s26 × s28 | 2 | 0 | 7 |
| 8 | s63 × s28 | 1 | 1 | 8 |
| 9 | ng124 × s16 | 2 | 0 | 7 |
| 10 | ng124 × s26 | 2 | 1 | 7 |
| 11 | ng124 × s34 | 0 | 0 | 3 |
| 12 | ng124 × s63 | 1 | 4 | 8 |
| 13 | ng124 × s44 | 2 | 0 | 5 |
. | Tetrad type . | |||
|---|---|---|---|---|
| Line . | Cross . | PD . | NPD . | T . |
| 1 | s34 × s26 | 41 | 0 | 0 |
| 2 | s34 × s63 | 43 | 0 | 0 |
| 3 | s44 × s28 | 37 | 0 | 0 |
| 4 | s28 × rgf1-Δ1a | 20 | 0 | 0 |
| 5 | s34 × s28 | 1 | 5 | 4 |
| 6 | s34 × s44 | 1 | 0 | 6 |
| 7 | s26 × s28 | 2 | 0 | 7 |
| 8 | s63 × s28 | 1 | 1 | 8 |
| 9 | ng124 × s16 | 2 | 0 | 7 |
| 10 | ng124 × s26 | 2 | 1 | 7 |
| 11 | ng124 × s34 | 0 | 0 | 3 |
| 12 | ng124 × s63 | 1 | 4 | 8 |
| 13 | ng124 × s44 | 2 | 0 | 5 |
Appropriate single-mutant strains (Table 1) were crossed, and tetrads were dissected and incubated at 30° or 32°. For crosses involving s44, spores were germinated on YE5S + 1 m sorbitol to reduce cell lysis. Mutant and wild-type segregants were distinguished by growing on YE5S medium containing phloxin B or (for ng124) by checking the morphological phenotypes microscopically.
Strain JW423 (Table 1).
Genetic interaction between the septins and Cdc4:
The s16 81nmt1-spn1+ 81nmt1-spn4+ triple mutant was viable at temperatures ranging from 23 to 36°. It seemed possible that the synthetic phenotype was ameliorated by residual septin expression from the nmt1 promoter under repressing conditions. Indeed, when s16 single-mutant (JW399) and spn4Δ (JW295) strains were crossed, 23 of 23 predicted double mutants were inviable and arrested as one or several highly elongated cells when tetrads were dissected on YE5S plates at 25° (data not shown). At 32°, 18 of 23 predicted double mutants were viable. Each inviable segregant arrested as a single, highly elongated cell, and most cells of a viable strain (JW401) had detectable defects in cytokinesis and/or septation. At 23°, although both single mutants grew at nearly normal rates, the double mutant grew very slowly (Figure 1E), and although some cells of the s16 single mutant formed partial or aberrant-looking septa (Figure 1I), the phenotype of the double mutant was much more severe: nearly all cells were highly elongated and had no or aberrant septa, and some cells were lysed (Figure 1M).
To identify the gene containing s16, we isolated several complementing plasmids, all of which contained cdc4 (see materials and methods). Moreover, when strains JW399 (s16) and JW401 (s16 spn4Δ) were transformed with plasmid pREP42∷GFP-cdc4, which expresses GFP-tagged Cdc4p from the medium-strength 41nmt1 promoter, they displayed wild-type and spn4Δ-like phenotypes, respectively, under inducing conditions (data not shown). In addition, s16 and the temperature-sensitive cdc4-8 allele are tightly linked. When JW399 was crossed to strain JW21 (cdc4-8), no wild-type segregants were detected after dissecting 19 tetrads (incubating segregants at 30°) and analyzing 468 random spores (half germinated at 32° and half at 25°). Finally, sequencing cdc4 from an s16 strain (see materials and methods) revealed a single G-to-A substitution in codon 36 that would result in the replacement of a glycine (conserved among myosin light chains) with glutamic acid. Because cdc4 is an essential gene (McCollum et al. 1995; Naqvi et al. 1999), the cdc4-s16 allele is presumably a hypomorphic or neomorphic allele that causes only a partial loss of Cdc4 function.
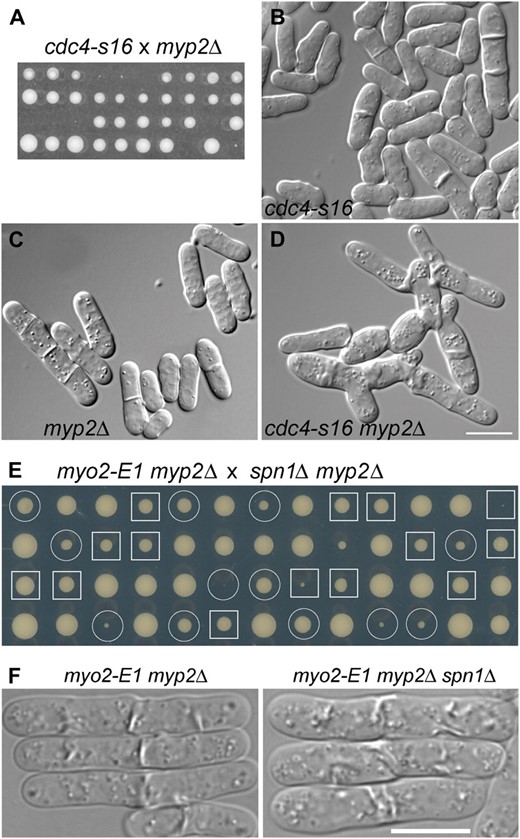
Among other binding partners (see discussion), Cdc4 is thought to be a common light chain for both type II myosin heavy chains, Myo2 and Myp2. Consistent with this model, we observed strong synthetic phenotypes between cdc4-s16 and both myp2 and myo2 mutations. When cdc4-s16 and myp2Δ strains were crossed, eight of nine predicted double mutants died or grew very poorly (Figure 2A), and only one grew well enough to culture further. At 30°, the single mutants had only mild cell-division defects (Figure 2, B and C), but the double mutant was severely defective in cytokinesis and septation (Figure 2D). Similarly, when strains JW400 (cdc4-s16) and YDM74 (myo2-E1, a temperature-sensitive allele) were crossed, all nine predicted double mutants from tetrads dissected on YE5S medium at 25° died as highly elongated cells (data not shown).
Genetic interactions among AMR and septin mutations. (A–D) Synthetic lethality between cdc4-s16 and myp2Δ. (A) Strains JW400 (cdc4-s16) and TP5 (myp2Δ) were crossed, and tetrads were dissected and incubated on YE5S medium at 32° for 6 days. Nine tetrads were tetratypes (i.e., contained one wild-type spore, two single-mutant spores, and one inviable or very slow-growing double-mutant spore) and one was a parental ditype (i.e., each spore was a single mutant and thus grew well). (B–D) DIC micrographs of aberrant cytokinesis in strains JW400, TP5, and JW345 (cdc4-s16 myp2Δ). Cells were grown overnight in YE5S liquid medium at 30°. Bar (B–D), 10 μm. (E and F) Absence of synthetic interaction between myo2-E1 myp2Δ and spn1Δ. (E) Strains TP90 (myo2-E1 myp2Δ) and JW380 (myp2Δ spn1Δ) were crossed, and tetrads were dissected and incubated on a YE5S plate at 25° for 7 days. Circles, myo2-E1 myp2Δ segregants; squares, myo2-E1 myp2Δ spn1Δ segregants. (F) Cells of strains TP90 and JW2560 were grown exponentially in YE5S liquid medium at 25°, shifted to 36° for 4 hr, and examined. Bar, 10 μm.
The septins localize to the division site ∼10 min before the initiation of septum formation (Berlin et al. 2003; Tasto et al. 2003; Wu et al. 2003), which is considerably later than Myo2 but similar to Myp2 (Kitayama et al. 1997; Bezanilla et al. 2000; Wu et al. 2003). Thus, it seemed possible that the septins might play a role in Myp2 localization or function; if so, then the synthetic lethality between cdc4-s16 and septin mutations might have the same basis as that between cdc4-s16 and myp2Δ, namely a partial loss of Myo2 function (due to the cdc4-s16 mutation) that would make full Myp2 function essential. To investigate this possibility, we used myo2-E1 to reduce Myo2 function and generated myo2-E1 spnΔ double mutants by crossing strain YDM74 to JW289 and JW295. However, although myo2-E1 and myp2Δ show a synthetic phenotype at all temperatures tested and are synthetically lethal at 34–36° (Bezanilla and Pollard 2000; Motegi et al. 2000), the myo2-E1 spnΔ strains displayed no obvious synthetic phenotypes at 25–36°. In addition, Myp2 localization appeared normal in an spn1Δ strain (Wu et al. 2003). Similarly, myp2Δ spn1Δ double mutants (from a cross of strains TP5 and JW289) displayed no obvious synthetic phenotypes at 18–36°, suggesting that the lack of septins does not significantly compromise the function of Myo2. Even a myo2-E1 myp2Δ spn1Δ triple mutant showed no synthetic growth defect at 23–32° (Figure 2E), and the severity of its cytokinesis defect was similar to that of the myo2-E1 myp2Δ double mutant (Figure 2F). Finally, although a wide variety of mutations with mild effects on the AMR become lethal when combined with a clp1Δ protein–phosphatase mutation (Mishra et al. 2004), an spn1Δ clp1Δ double mutant displayed no detectable synthetic effect at 18–36° (data not shown). Thus, the synthetic lethality between cdc4-s16 and spn1Δ or spn4Δ does not appear to result from synergistic effects of the cdc4 and spn mutations on the function of the AMR and presumably has some other basis (see discussion).
Interaction between the septins and a cell-wall-integrity pathway:
As described above, mutations s34, s26, and s63 mapped to one locus and s44 and s28 to another. These mutants all had a similar phenotype that was strongest in s34 and s44, which were therefore characterized further. Under nmt1-repressing conditions, the s34 and s44 single mutants and an 81nmt1-spn1+ 81nmt1-spn4+ strain all grew reasonably well, but the triple mutants grew very poorly (Figure 1, F and G), and 24 hr after a shift from inducing to repressing conditions, 5% of 670 s34 cells, 11% of 1218 s44 cells, 71% of 820 s34 81nmt1-spn1+ 81nmt1-spn4+ cells, and 76% of 779 s44 81nmt1-spn1+ 81nmt1-spn4+ cells appeared shrunken or lysed (Figure 1, J, K, N, and O). These phenotypes appeared similar at temperatures ranging from 18 to 36°. We also examined the phenotypes of s34 spnΔ and s44 spnΔ strains produced by crosses among strains JW403, JW289, JW295, and JW406. In each case, most double-mutant segregants arrested as a single elongated cell, while some formed microcolonies with many lysed cells. Inclusion of 1.2 m sorbitol in the medium partially rescued the phenotype and allowed the recovery of viable s34 spnΔ and s44 spnΔ strains.
To examine the cell-lysis phenotype more closely, we performed time-lapse microscopy on an s34 spn1Δ strain (Figure 3A; supporting information, File S1). When s34 81nmt1-spn1+ 81nmt1-spn4+ (JW402) and s44 81nmt1-spn1+ 81nmt1-spn4+(W405) strains were grown under repressing conditions, they gave very similar results. In the starting populations, some cells had already lysed (e.g., Figure 3A, time 0, lower right) and others were present as short chains of unseparated daughter cells (e.g., Figure 3A, time 0, cells a, b, and c), reflecting the delay in cell separation due to the lack of septins. The time-lapse observations suggested that cell lysis occurred primarily or exclusively during attempted cell separation and in several different patterns. In some cases, only one of the two separating cells lysed [Figure 3A, cell a (time 00:52:00) and cell b (time 00:14:00)]; in other cases, both daughter cells lysed simultaneously (Figure 3A, cell b, compare times 00:52:00 and 00:52:30) or sequentially within ∼30 sec (Figure 3A, cell c, time 01:09:00 to time 01:10:30). Interestingly, in some cases, a cell lying between two lysed cells remained intact throughout the entire period of observation (Figure 3A, cell b, second compartment from the left). Although most cells lysed when they attempted to separate, some cells separated successfully, but slowly (>110 min after the appearance of the septum, in contrast to ∼50–60 min for the septation-to-separation interval in wild-type cells) (Figure 3A, cell a, right-hand cells, 01:48:00). During the following 5 hr, the daughter cells that had not lysed continued to grow and formed new septa without lysis, supporting the hypothesis that lysis occurred only at the time of attempted cell separation.
Cell-lysis phenotype of the s34 spn1 double mutant and normal septin localization in the s34 and s44 mutants. (A) Lysis during cell separation in s34 spn1Δ strain JW374. Cells were grown to exponential phase in EMM5S + 1 m sorbitol at 30°, washed twice in EMM5S, and observed by time-lapse DIC photomicroscopy on EMM5S + 25% gelatin at 24° (see materials and methods). Selected images are shown from a series recorded at 30-sec intervals; times are indicated in hours, minutes, and seconds. Cells are labeled for reference in the text. The entire series can be viewed in File S1. (B) Localization of Spn1 in s34 spn1-mEGFP strain JW1126 (left panels) and s44 spn1-mEGFP strain JW1125 (right panels). Cells were grown in YE5S at 25° and observed by DIC and fluorescence microscopy. Arrowheads in B, lysed cells. Bars, 10 μm.
Although a few percent of cells harboring only septin mutations also lyse (Figure 1, C and D; Table 3), the strong synthetic phenotypes observed with s34 and s44 suggested that the phenotypes due to these mutations did not result from an interference with septin localization or function. Consistent with this interpretation, it appeared that the septins localized normally to the division site in s34 and s44 mutant cells (Figure 3B).
Accumulation of cells with abnormal numbers and/or structures of septa in various mutant strains
Strain . | No. of cells scored . | % of cells with the indicated number of septaa . | % of lysed or shrunken cellsb . | % of cells with thick walls or septac . | ||||
|---|---|---|---|---|---|---|---|---|
| 0 . | 1 . | 2 . | 3 . | ≥4 . | ||||
| Wild type | 434 | 86 | 14 | 0 | 0 | 0 | 0 | 0 |
| spn4Δ | 487 | 29 | 56 | 5 | 3 | 0 | 7 | 0 |
| 3nmt1-rgf1 | 434 | 45 | 50 | 4 | 1 | 0 | 0 | 16d |
| 3nmt1-rgf2 | 400 | 39 | 44 | 9 | 6 | 3 | 0 | 73d |
| 3nmt1-rgf3 | 363 | 33 | 53 | 9 | 4 | 1 | 0 | 5d |
| ng124 | 688 | 58 | 31 | 8 | 2 | 1 | 1 | 0 |
| ng124 spn1Δ | 564 | 3 | 32 | 16 | 23 | 16 | 17d + 10 | 0 |
| mid2Δ | 400 | 37 | 58 | 1 | 2 | 0 | 2 | 0 |
| mid2Δ spn4Δ | 465 | 52 | 44 | 1 | 1 | 0 | 2 | 0 |
| ng124 mid2Δ | 613 | 3 | 52 | 13 | 29 | 3 | 17d | 0 |
| spn1Δ | 1240 | 45 | 50 | 2 | 3 | 0 | ND | ND |
| scw1Δ | 1570 | 54 | 40 | 3 | 2 | 0.1 | ND | ND |
| scw1Δ spn1Δ | 1237 | 7 | 47 | 14 | 27 | 5 | ND | ND |
Strain . | No. of cells scored . | % of cells with the indicated number of septaa . | % of lysed or shrunken cellsb . | % of cells with thick walls or septac . | ||||
|---|---|---|---|---|---|---|---|---|
| 0 . | 1 . | 2 . | 3 . | ≥4 . | ||||
| Wild type | 434 | 86 | 14 | 0 | 0 | 0 | 0 | 0 |
| spn4Δ | 487 | 29 | 56 | 5 | 3 | 0 | 7 | 0 |
| 3nmt1-rgf1 | 434 | 45 | 50 | 4 | 1 | 0 | 0 | 16d |
| 3nmt1-rgf2 | 400 | 39 | 44 | 9 | 6 | 3 | 0 | 73d |
| 3nmt1-rgf3 | 363 | 33 | 53 | 9 | 4 | 1 | 0 | 5d |
| ng124 | 688 | 58 | 31 | 8 | 2 | 1 | 1 | 0 |
| ng124 spn1Δ | 564 | 3 | 32 | 16 | 23 | 16 | 17d + 10 | 0 |
| mid2Δ | 400 | 37 | 58 | 1 | 2 | 0 | 2 | 0 |
| mid2Δ spn4Δ | 465 | 52 | 44 | 1 | 1 | 0 | 2 | 0 |
| ng124 mid2Δ | 613 | 3 | 52 | 13 | 29 | 3 | 17d | 0 |
| spn1Δ | 1240 | 45 | 50 | 2 | 3 | 0 | ND | ND |
| scw1Δ | 1570 | 54 | 40 | 3 | 2 | 0.1 | ND | ND |
| scw1Δ spn1Δ | 1237 | 7 | 47 | 14 | 27 | 5 | ND | ND |
Cultures were grown to exponential phase in EMM5S liquid medium at either 30° [strains JW81 (wild type), JW295 (spn4Δ), JW331 (ng124), JW321-1 (ng124 spn1Δ), JW430 (mid2Δ), JW314 (mid2Δ spn4Δ), and JW315 (ng124 mid2Δ)] or 25° [strains JW1078 (3nmt1-rgf1), JW903 (3nmt1-rgf2), JW1081 (3nmt1-rgf3), JW290 (spn1Δ), DM1274 (scw1Δ), and JW2286 (spn1Δ scw1Δ)] before staining with Calcofluor and examining by fluorescence microscopy. ND, not determined.
Some counts total <100% because the numbers of septa were not scored in the lysed or shrunken cells.
Whole cells or individual compartments of septated cells appeared to be lysed.
Cells with very bright Calcofluor staining compared to wild-type cells.
In these cases, the numbers of septa in the cells were also scored.
Accumulation of cells with abnormal numbers and/or structures of septa in various mutant strains
Strain . | No. of cells scored . | % of cells with the indicated number of septaa . | % of lysed or shrunken cellsb . | % of cells with thick walls or septac . | ||||
|---|---|---|---|---|---|---|---|---|
| 0 . | 1 . | 2 . | 3 . | ≥4 . | ||||
| Wild type | 434 | 86 | 14 | 0 | 0 | 0 | 0 | 0 |
| spn4Δ | 487 | 29 | 56 | 5 | 3 | 0 | 7 | 0 |
| 3nmt1-rgf1 | 434 | 45 | 50 | 4 | 1 | 0 | 0 | 16d |
| 3nmt1-rgf2 | 400 | 39 | 44 | 9 | 6 | 3 | 0 | 73d |
| 3nmt1-rgf3 | 363 | 33 | 53 | 9 | 4 | 1 | 0 | 5d |
| ng124 | 688 | 58 | 31 | 8 | 2 | 1 | 1 | 0 |
| ng124 spn1Δ | 564 | 3 | 32 | 16 | 23 | 16 | 17d + 10 | 0 |
| mid2Δ | 400 | 37 | 58 | 1 | 2 | 0 | 2 | 0 |
| mid2Δ spn4Δ | 465 | 52 | 44 | 1 | 1 | 0 | 2 | 0 |
| ng124 mid2Δ | 613 | 3 | 52 | 13 | 29 | 3 | 17d | 0 |
| spn1Δ | 1240 | 45 | 50 | 2 | 3 | 0 | ND | ND |
| scw1Δ | 1570 | 54 | 40 | 3 | 2 | 0.1 | ND | ND |
| scw1Δ spn1Δ | 1237 | 7 | 47 | 14 | 27 | 5 | ND | ND |
Strain . | No. of cells scored . | % of cells with the indicated number of septaa . | % of lysed or shrunken cellsb . | % of cells with thick walls or septac . | ||||
|---|---|---|---|---|---|---|---|---|
| 0 . | 1 . | 2 . | 3 . | ≥4 . | ||||
| Wild type | 434 | 86 | 14 | 0 | 0 | 0 | 0 | 0 |
| spn4Δ | 487 | 29 | 56 | 5 | 3 | 0 | 7 | 0 |
| 3nmt1-rgf1 | 434 | 45 | 50 | 4 | 1 | 0 | 0 | 16d |
| 3nmt1-rgf2 | 400 | 39 | 44 | 9 | 6 | 3 | 0 | 73d |
| 3nmt1-rgf3 | 363 | 33 | 53 | 9 | 4 | 1 | 0 | 5d |
| ng124 | 688 | 58 | 31 | 8 | 2 | 1 | 1 | 0 |
| ng124 spn1Δ | 564 | 3 | 32 | 16 | 23 | 16 | 17d + 10 | 0 |
| mid2Δ | 400 | 37 | 58 | 1 | 2 | 0 | 2 | 0 |
| mid2Δ spn4Δ | 465 | 52 | 44 | 1 | 1 | 0 | 2 | 0 |
| ng124 mid2Δ | 613 | 3 | 52 | 13 | 29 | 3 | 17d | 0 |
| spn1Δ | 1240 | 45 | 50 | 2 | 3 | 0 | ND | ND |
| scw1Δ | 1570 | 54 | 40 | 3 | 2 | 0.1 | ND | ND |
| scw1Δ spn1Δ | 1237 | 7 | 47 | 14 | 27 | 5 | ND | ND |
Cultures were grown to exponential phase in EMM5S liquid medium at either 30° [strains JW81 (wild type), JW295 (spn4Δ), JW331 (ng124), JW321-1 (ng124 spn1Δ), JW430 (mid2Δ), JW314 (mid2Δ spn4Δ), and JW315 (ng124 mid2Δ)] or 25° [strains JW1078 (3nmt1-rgf1), JW903 (3nmt1-rgf2), JW1081 (3nmt1-rgf3), JW290 (spn1Δ), DM1274 (scw1Δ), and JW2286 (spn1Δ scw1Δ)] before staining with Calcofluor and examining by fluorescence microscopy. ND, not determined.
Some counts total <100% because the numbers of septa were not scored in the lysed or shrunken cells.
Whole cells or individual compartments of septated cells appeared to be lysed.
Cells with very bright Calcofluor staining compared to wild-type cells.
In these cases, the numbers of septa in the cells were also scored.
To identify the genes containing s34 and s44, we first screened genomic DNA libraries for plasmids that could rescue the phenotypes of s34 spn1 spn4 and s44 spn1 spn4 strains. With s34, we recovered 13 plasmids whose inserts were mapped to six distinct chromosomal locations by restriction digestion and partial sequencing (Table 4). With s44, we recovered 23 plasmids whose inserts mapped to three distinct chromosomal locations that were a subset of those isolated with s34 (Table 4). Remarkably, the genes contained in these plasmids all appear to represent components of an S. pombe pathway for the maintenance of cell-wall integrity (see discussion).
Rescue of mutations s34 and s44 by plasmid-borne genes
Mutant . | Chromosome regiona . | No. of plasmids . | Presumed relevant gene(s) . |
|---|---|---|---|
| s34 | IIL (C30B4 and C3D6) | 1 | wsc1 (homolog of S. cerevisiae WSC genes, which encode sensors of surface stress)b |
| IR (C1006) | 1 | rgf2 (encodes a Rho-GEF)c | |
| IIICEN (C645) | 2 | rgf1 and rgf3 (encode Rho-GEFs)d | |
| IR (C1F7) | 4 | rho1e | |
| IIR (C1289) | 3 | pob1 (encodes homolog of S. cerevisiae Boi1 and Boi2, involved in Rho protein signal transduction)f | |
| IIR (C12D12) | 2 | pck2 (encodes a protein kinase C)g | |
| s44 | IIL (C30B4 and C3D6) | 1 | wsc1 (see above)h |
| IIICEN (C645) | 3 | rgf1 and rgf3 (see above)i | |
| IR (C1F7) | 19 | rho1j |
Mutant . | Chromosome regiona . | No. of plasmids . | Presumed relevant gene(s) . |
|---|---|---|---|
| s34 | IIL (C30B4 and C3D6) | 1 | wsc1 (homolog of S. cerevisiae WSC genes, which encode sensors of surface stress)b |
| IR (C1006) | 1 | rgf2 (encodes a Rho-GEF)c | |
| IIICEN (C645) | 2 | rgf1 and rgf3 (encode Rho-GEFs)d | |
| IR (C1F7) | 4 | rho1e | |
| IIR (C1289) | 3 | pob1 (encodes homolog of S. cerevisiae Boi1 and Boi2, involved in Rho protein signal transduction)f | |
| IIR (C12D12) | 2 | pck2 (encodes a protein kinase C)g | |
| s44 | IIL (C30B4 and C3D6) | 1 | wsc1 (see above)h |
| IIICEN (C645) | 3 | rgf1 and rgf3 (see above)i | |
| IR (C1F7) | 19 | rho1j |
Plasmids were recovered during attempts to clone the genes containing mutations s34 and s44 (see materials and methods).
The chromosome arm and relevant cosmid(s) from the S. pombe genome project are indicated.
The plasmid contains the entire 1125-bp (no introns) ORF SPBC30B4.01c [designated wsc1 on the basis of the similarity of its product to the S. cerevisiae Wsc proteins (Levin 2005)] plus ∼2.1 kb of downstream flanking sequence; the 1326 bp between wsc1 and the uncharacterized ORF SPBC30B4.02c; and the C-terminal 822 bp (∼39%) of SPBC30B4.02c, which seems unlikely to be relevant. The 2.1 kb downstream of wsc1 contains no known or predicted genes.
The plasmid contains the entire 3576-bp coding region (including two introns totaling 99 bp) of rgf2 (SPAC1006.06); the 729 bp between rgf2 and the divergently transcribed och1 (SPAC1006.5c); the entire 1191-bp (no introns) ORF of och1 (encoding an α-1,6-mannosyltransferase) plus 314 bp of downstream flanking sequence; the 883 bp between rgf2 and ORF SPAC1006.07; and the N-terminal 337 bp (∼29%) of this ORF (which encodes a putative translation factor). och1 and SPAC1006.07 seem unlikely to be relevant.
One plasmid contains the entire 3828-bp (no introns) rgf3 (SPCC645.06c) plus ∼850 bp of upstream sequence; the 1355 bp between rgf3 and myo2 (SPCC645.05c); the N-terminal 2307 bp of the 4581-bp (no introns) myo2 ORF; but no part of rgf1 (SPCC645.07). The other plasmid contains the N-terminal 1229 bp (∼32%) of rgf3 (which does not include the Rho-GEF domain); the 1051 bp between rgf3 and the divergently transcribed rgf1; and the N-terminal 3959 bp of the 4051-bp coding region (including one 46-bp intron) of rgf1.
Each plasmid has one or the other of two distinct inserts, each of which contains the entire 940-bp coding region (including one 331-bp intron) of rho1 (SPAC1F7.04) plus flanking sequences. The region of overlap between the inserts contains no other known or predicted genes.
The plasmids have three distinct inserts, each of which contains the entire 2811-bp coding region (including one 195-bp intron) of pob1 (SPBC1289.04c) plus flanking sequences. The region of overlap between the inserts contains no other known or predicted genes.
The region shared by the two plasmid inserts contains the entire 3051-bp (no introns) ORF of pck2 (SPBC12D12.04c); the entire 933-bp coding region (including five introns totaling 291 bp) of rev7 (SPBC12D12.09, encoding a putative DNA polymerase ζ); and the C-terminal 43 bp (∼3%) and 306 bp (∼24%) of cct1 (SPBC12D12.03) and SPBC12D12.05c, respectively, which seem unlikely to be relevant. [Note that the SPBC12D12.09 ORF number is out of sequence because this gene was missed during initial annotation (V. Wood, personal communication).]
The plasmid contains the C-terminal 797 bp (∼71%) of the wsc1 ORF (see footnote b) plus 1584 bp of downstream sequence that contains no other known or predicted gene.
The three plasmids have identical inserts that contain only an internal 1930-bp fragment (lacking the N-terminal 1681 bp and the C-terminal 440 bp) of the rgf1 coding sequence (see footnote d). This fragment contains the Rho-GEF domain. In addition, the two rgf1/rgf3 plasmids isolated in attempting to clone the s34 gene (see footnote d) were also able to rescue the phenotype of the s44 strain JW405.
The plasmids have at least six distinct inserts, all of which contain the entire rho1 gene (see footnote e) plus flanking sequences. The region of overlap between the inserts contains no other known or predicted genes.
Rescue of mutations s34 and s44 by plasmid-borne genes
Mutant . | Chromosome regiona . | No. of plasmids . | Presumed relevant gene(s) . |
|---|---|---|---|
| s34 | IIL (C30B4 and C3D6) | 1 | wsc1 (homolog of S. cerevisiae WSC genes, which encode sensors of surface stress)b |
| IR (C1006) | 1 | rgf2 (encodes a Rho-GEF)c | |
| IIICEN (C645) | 2 | rgf1 and rgf3 (encode Rho-GEFs)d | |
| IR (C1F7) | 4 | rho1e | |
| IIR (C1289) | 3 | pob1 (encodes homolog of S. cerevisiae Boi1 and Boi2, involved in Rho protein signal transduction)f | |
| IIR (C12D12) | 2 | pck2 (encodes a protein kinase C)g | |
| s44 | IIL (C30B4 and C3D6) | 1 | wsc1 (see above)h |
| IIICEN (C645) | 3 | rgf1 and rgf3 (see above)i | |
| IR (C1F7) | 19 | rho1j |
Mutant . | Chromosome regiona . | No. of plasmids . | Presumed relevant gene(s) . |
|---|---|---|---|
| s34 | IIL (C30B4 and C3D6) | 1 | wsc1 (homolog of S. cerevisiae WSC genes, which encode sensors of surface stress)b |
| IR (C1006) | 1 | rgf2 (encodes a Rho-GEF)c | |
| IIICEN (C645) | 2 | rgf1 and rgf3 (encode Rho-GEFs)d | |
| IR (C1F7) | 4 | rho1e | |
| IIR (C1289) | 3 | pob1 (encodes homolog of S. cerevisiae Boi1 and Boi2, involved in Rho protein signal transduction)f | |
| IIR (C12D12) | 2 | pck2 (encodes a protein kinase C)g | |
| s44 | IIL (C30B4 and C3D6) | 1 | wsc1 (see above)h |
| IIICEN (C645) | 3 | rgf1 and rgf3 (see above)i | |
| IR (C1F7) | 19 | rho1j |
Plasmids were recovered during attempts to clone the genes containing mutations s34 and s44 (see materials and methods).
The chromosome arm and relevant cosmid(s) from the S. pombe genome project are indicated.
The plasmid contains the entire 1125-bp (no introns) ORF SPBC30B4.01c [designated wsc1 on the basis of the similarity of its product to the S. cerevisiae Wsc proteins (Levin 2005)] plus ∼2.1 kb of downstream flanking sequence; the 1326 bp between wsc1 and the uncharacterized ORF SPBC30B4.02c; and the C-terminal 822 bp (∼39%) of SPBC30B4.02c, which seems unlikely to be relevant. The 2.1 kb downstream of wsc1 contains no known or predicted genes.
The plasmid contains the entire 3576-bp coding region (including two introns totaling 99 bp) of rgf2 (SPAC1006.06); the 729 bp between rgf2 and the divergently transcribed och1 (SPAC1006.5c); the entire 1191-bp (no introns) ORF of och1 (encoding an α-1,6-mannosyltransferase) plus 314 bp of downstream flanking sequence; the 883 bp between rgf2 and ORF SPAC1006.07; and the N-terminal 337 bp (∼29%) of this ORF (which encodes a putative translation factor). och1 and SPAC1006.07 seem unlikely to be relevant.
One plasmid contains the entire 3828-bp (no introns) rgf3 (SPCC645.06c) plus ∼850 bp of upstream sequence; the 1355 bp between rgf3 and myo2 (SPCC645.05c); the N-terminal 2307 bp of the 4581-bp (no introns) myo2 ORF; but no part of rgf1 (SPCC645.07). The other plasmid contains the N-terminal 1229 bp (∼32%) of rgf3 (which does not include the Rho-GEF domain); the 1051 bp between rgf3 and the divergently transcribed rgf1; and the N-terminal 3959 bp of the 4051-bp coding region (including one 46-bp intron) of rgf1.
Each plasmid has one or the other of two distinct inserts, each of which contains the entire 940-bp coding region (including one 331-bp intron) of rho1 (SPAC1F7.04) plus flanking sequences. The region of overlap between the inserts contains no other known or predicted genes.
The plasmids have three distinct inserts, each of which contains the entire 2811-bp coding region (including one 195-bp intron) of pob1 (SPBC1289.04c) plus flanking sequences. The region of overlap between the inserts contains no other known or predicted genes.
The region shared by the two plasmid inserts contains the entire 3051-bp (no introns) ORF of pck2 (SPBC12D12.04c); the entire 933-bp coding region (including five introns totaling 291 bp) of rev7 (SPBC12D12.09, encoding a putative DNA polymerase ζ); and the C-terminal 43 bp (∼3%) and 306 bp (∼24%) of cct1 (SPBC12D12.03) and SPBC12D12.05c, respectively, which seem unlikely to be relevant. [Note that the SPBC12D12.09 ORF number is out of sequence because this gene was missed during initial annotation (V. Wood, personal communication).]
The plasmid contains the C-terminal 797 bp (∼71%) of the wsc1 ORF (see footnote b) plus 1584 bp of downstream sequence that contains no other known or predicted gene.
The three plasmids have identical inserts that contain only an internal 1930-bp fragment (lacking the N-terminal 1681 bp and the C-terminal 440 bp) of the rgf1 coding sequence (see footnote d). This fragment contains the Rho-GEF domain. In addition, the two rgf1/rgf3 plasmids isolated in attempting to clone the s34 gene (see footnote d) were also able to rescue the phenotype of the s44 strain JW405.
The plasmids have at least six distinct inserts, all of which contain the entire rho1 gene (see footnote e) plus flanking sequences. The region of overlap between the inserts contains no other known or predicted genes.
Linkage analysis allowed identification of the genes actually containing s34 and s44. s34 showed linkage (∼20 cM) to the mat locus, suggesting that it might be an allele of the nearby bgs1, a plausible target (see Introduction) of the cell-wall-integrity pathway. Indeed, a cross of strains JW404 and MBY580 revealed tight linkage (∼1 cM) between the s34 and bgs1-191 mutations. However, sequencing of genomic DNA from an s34 strain revealed no mutations in the bgs1 ORF or its flanking regions. Additional sequencing revealed a single C-to-T substitution at nucleotide 133 of the 1449-bp ORF SPBC19G7.08c, producing a TGA (stop) codon and presumably a null allele. bgs1-191 has a mutation in codon 277 (Liu et al. 1999), 10.18 kb from the putative s34 mutation, which is consistent with the tight linkage observed. SPBC19G7.08c, now designated art1, encodes a previously unstudied arrestin-like protein. The phenotype of an art1Δ mutant closely resembles that of the s34 mutant, and Art1 localizes to the division site (R. Davidson, D. Laporte and J.-Q. Wu, unpublished results), which is consistent with its apparent role in cytokinesis. Further studies of the function of this protein are in progress.
In a cross of s44 (JW407) and myo2-E1 (YDM74) strains, no recombinants were observed between these markers in 49 tetrads. Because myo2 is adjacent to the tandem rgf3 and rgf1 (see Table 4), on the rgf3 side, these data suggested that s44 is an allele of rgf3. However, plasmids containing either rgf3 or rgf1 alone can rescue the s44 mutation (Table 4, footnotes d and i), and a cross between s28 and rgf1Δ strains also yielded no wild-type recombinants in 20 tetrads (Table 2, line 4). Sequencing genomic DNA from an s44 strain revealed a single C-to-T substitution in codon 517 of rgf3, resulting in a G-to-E substitution in the Rho-GEF domain. Thus, s44 is an allele of rgf3, and s28 could be in either rgf3 or rgf1.
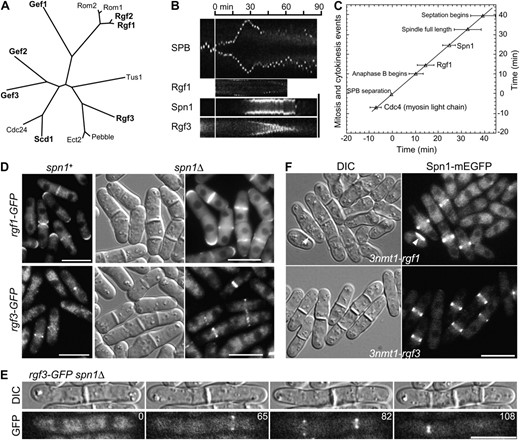
Independence of septin and Rho-GEF localization to the division site:
As described above, mutation s44 lies in rgf3, and rgf1+, rgf2+, and rgf3+, along with rho1+, were all recovered as dosage suppressors of s44 and/or of the phenotypically similar mutation s34. The rgf genes encode members of the Rho-GEF family (Figure 4A) and have been studied extensively in the past few years (Iwaki et al. 2003; Tajadura et al. 2004; Morrell-Falvey et al. 2005; Mutoh et al. 2005; García et al. 2006, 2009a,b); all three proteins appear to have Rho1 as a primary or exclusive target. In agreement with these other studies, we found that all three proteins localized to the division site at around the time of cell division (Figure 4, B–D; note that Rgf2, which required overexpression to be visualized, was not studied in as much detail). In addition, Rgf1, but not Rgf2 or Rgf3, localized to a cap at one or both cell poles during interphase (Figure 4D).
Independence of septin and Rho-GEF localization to the division site. (A) Relationships among Rho-GEFs from S. pombe (http://www.genedb.org/genedb/pombe/) and S. cerevisiae (http://www.yeastgenome.org/), as well as a Rho-GEF from Drosophila (Pebble) and one from humans (ECT2) that have also been implicated in cytokinesis (Miki et al. 1993; Prokopenko et al. 1999; Tatsumoto et al. 1999; O'Keefe et al. 2001; Giansanti et al. 2004; Shandala et al. 2004). Clustal W was used to align the Rho-GEF domains and then derive a phylogenetic tree based on this alignment. (B and C) Localization and dynamic behavior of Rgf1 and Rgf3 at the division site in relation to the septins and other cell-division markers in wild-type cells. Strains JW1113 (spn1-mEGFP sad1-mRFP1), JW1124 (rgf1-GFP sad1-mRFP1), and JW1131 (rgf3-mEGFP sad1-mRFP1) were filmed in EMM5S at 25°. (B) Kymographs constructed using Image J and the movements of the spindle-pole bodies (SPBs), as marked by Sad1-mRFP1, to allow alignment at the time of SPB separation (vertical line). For the SPB, a 13.7-μm slit parallel to the long axis of the cell was used; for the other proteins, a 4.1-μm slit across the midplane of the cell was used. (C) Timeline showing the initial appearance of Spn1 (n = 8 cells) and Rgf1 (n = 8) at the division site. SPB separation is defined as time zero, and the mean time of appearance of each protein (±1 standard deviation) is plotted. The beginning of septation was scored by DIC. The data for Cdc4 are from Wu et al. (2003). (D and E) Tests of possible septin dependence of Rho-GEF localization. (D) Strains JW1124 (rgf1-GFP), JW1139 (rgf1-GFP spn1Δ), JW1131 (rgf3-mEGFP), and JW1128 (rgf3-GFP spn1Δ) were grown in EMM5S (JW1124 and JW1131) or YE5S (JW1139 and JW1128) medium at 25° and examined by DIC and/or fluorescence microscopy. (E) Strain JW1128 was examined by time-lapse microscopy. Selected DIC and GFP images (elapsed times given in minutes) are shown from a series recorded at 1-min intervals. The entire series can be viewed in File S2. (F) Cell morphology and septin localization in cells overexpressing Rgf1 or Rgf3. Strains JW1123 (spn1-mEGFP 3nmt1-rgf1) and JW1122 (spn1-mEGFP 3nmt1-rgf3) were grown under inducing conditions (EMM5S medium) for 24 hr at 25°; paired DIC and fluorescence images are shown. Arrowheads indicate a cell with a region of thickened cell wall. Bars (B and D–F), 10 μm.
Because the septins also assemble at the division site at about the same time as the Rgf proteins, it seemed possible that localization of the Rgf proteins to the division site would depend on a septin scaffold (see Introduction). However, Rgf1 and Rgf3 appeared to localize normally to the division site at the appropriate times in a septin mutant (Figure 4, D and E; File S2). In addition, the Rgf1 and Rgf3 rings constricted as the furrow ingressed during cytokinesis in wild-type cells, whereas the septin rings did not (Figure 4B).
In further agreement with the published studies, we found that overexpression of the Rgf proteins produced cells with multiple and/or aberrant septa (Figure 4F; Table 3). Because this phenotype resembled that of septin mutants, it seemed possible that overexpression of the Rgf proteins interfered with septin localization. However, Spn1 appeared to localize normally when either Rgf1 or Rgf3 was overexpressed (Figure 4F). In addition, the mild vegetative phenotypes of rgf1Δ and rgf2Δ mutants (Morrell-Falvey et al. 2005; Mutoh et al. 2005; García et al. 2006, 2009b; our unpublished data) suggest that septin function is not perturbed by a lack of Rgf1 or Rgf2, and septins also appeared to localize normally in an rgf3-s44 mutant strain (Figure 3B).
Taken together, the data suggest that the pathway involving Rho1 and its GEFs functions independently of that involving the septins, although the two pathways may complement each other in function.
A mutation that may identify a pathway parallel to that of the septins:
The original ng124 81nmt1-spn1+ 81nmt1-spn4+ triple mutant had a subtle growth defect but a conspicuous cell-separation delay. After backcrossing, ng124 single mutants grew well (Figure 1H) but had a mild cell-separation delay like that of septin mutants (Figure 1L; Figure 5B; Table 3). An ng124 spn1Δ double mutant also grew reasonably well (Figure 1H) but displayed a much more severe cell-separation delay, such that nearly all cells contained one or more septa, and some cells had lysed (Figure 1P; Table 3). This phenotype was similar at temperatures ranging from 18° to 36° but was more dramatic on solid media (where many cells had up to seven or nine septa) than in liquid media. In a typical time-lapse series (Figure 5A; File S3), no successful cell separation was observed during >4.5 hr of filming, although some older septa became very abnormal in shape (Figure 5A, arrows) and some cells lysed (Figure 5A, arrowheads; Table 3).
Characterization of mutant ng124 and its identification as an allele of scw1. (A and B) Selected DIC and fluorescence images are shown from time-lapse-microscopy series recorded at 1- or 2-min intervals; times are indicated in hours and minutes. The entire series can be viewed in File S3 and File S4. (A) Cell-separation defect in an ng124 spn1Δ double mutant. Strain JW321-1 was grown in YE5S medium at 30° and observed on YE5S at 24°. Arrows indicate abnormal-shaped septa; arrowheads indicate cells that appear to have lysed. (B) Apparently normal septin localization and variable cell-separation delay in an ng124 single mutant. Strain JW320 (ng124 spn4-GFP) was grown in EMM5S at 23° and observed on EMM5S at 24°. Cells are numbered for reference in the text. (C) Noncomplementation of ng124 and scw1Δ in diploid cells. Diploid strains JW2613 (scw1+/ng124) and JW2614 (scw1Δ/ng124) were observed by DIC microscopy after growth on YE5S-Ade at 25°. (D) Rescue of ng124 by plasmids containing scw1+. Strain JW2155 (ng124) was transformed with empty vector or with chromosome III plasmids that contain only scw1 (nucleotides 673,476–675,161) in their region of overlap (nucleotides 672,482–675,308), grown in EMM5S-Leu medium at 25°, and observed by DIC microscopy. Bars, 10 μm.
Although the ng124 single-mutant phenotype resembled that of septin mutants, the synthetic phenotype suggested that ng124 did not interfere with septin localization or function. Consistent with this interpretation, time-lapse and confocal microscopy showed that the septins appeared at the presumptive division site at the normal time (just before septum formation) and in the normal double-ring configuration in an ng124 mutant [Figure 5B, particularly cell 3 at 00:00 (note septin rings visible at two sites where septa are just beginning to form) and cell 1 at 01:02 and 01:17; Figure 6, A–C; File S4, File S5, and File S6]. In addition, although the times elapsing between septum formation and cell separation were highly variable (Figure 5B: compare cell 1 at 01:17, 02:33, and 02:36 to cells 3 and 4 at 00:00 and 02:36), the septins appeared to disassemble from the septation site at approximately the normal time (i.e., when cell separation would normally occur) regardless of whether or not cell separation occurred on schedule at that particular site (Figure 5B: compare cell 1 at 01:17, 02:33, and 02:36 to cells 3 and 4 at 00:00, 01:02, 01:17, and 02:33).
Functional relationships among the septins, Mid2, and Scw1. Cells were observed by spinning-disk confocal microscopy (A–F) or by wide-field fluorescence microscopy (G–J) after growth in YE5S at 25° (A–G) or EMM5S at 30° (H–J). In each case, the cells shown are representative of large numbers of cells examined. For the fluorescence images in A–F, stacks of 70 z-sections (0.07-μm) were captured using the same laser power and acquisition settings and projected into three-dimensional images using Image J (see materials and methods). Selected views of the three-dimensional images are shown with corresponding DIC images; the entire series can be viewed in File S5, File S6, File S7, File S8, and File S9. (A and B) Localization of Spn1-GFP in wild-type strain JW306. The images in B are a stereo pair. (C) Localization of Spn4-GFP in scw1-ng124 strain JW320. (D) Localization of Mid2-GFP in wild-type strain JW326. (E) Localization of Spn1-GFP in mid2Δ strain JW318. (F) Localization of Mid2-GFP in scw1-ng124 strain JW1203. (G) Localization of Mid2-GFP in spn4Δ strain JW332. (H–J) Mutant phenotypes of strains (H) JW430 (mid2Δ), (I) JW315 (scw1-ng124 mid2Δ), and (J) JW314 (spn4Δ mid2Δ). Cells were stained with bisbenzimide to view nuclei and septa. Bars, 10 μm.
mid2Δ mutants also have a cell-separation delay resembling that of septin mutants (Berlin et al. 2003; Tasto et al. 2003; Figure 6H; Table 3). Like ng124 spnΔ double mutants, an ng124 mid2Δ double mutant has a strong synthetic phenotype, with more frequent and longer chains of cells than found in the single mutants (Figure 6I; Table 3). In striking contrast, spn4Δ mid2Δ double mutants display no obvious synthetic phenotype and resemble the two single mutants (Berlin et al. 2003; Tasto et al. 2003; Figure 6J; Table 3). These observations suggest that Mid2 and the septins function together in one pathway while the product of the gene identified by ng124 functions in a distinct and perhaps redundant pathway. Observations of protein localization are consistent with this hypothesis. Mid2 and the septins colocalize interdependently to a double ring at the division site (Berlin et al. 2003; Tasto et al. 2003; Figure 6, A, B, D, E, and G; File S5, File S7, and File S8). In contrast, both the septins (see above) and Mid2 (Figure 6F; File S9) appear to localize almost normally in ng124 mutant strains.
After failing to identify the ng124-containing gene by plasmid rescue using the available libraries, we used positional cloning (see materials and methods), which suggested that ng124 might be an allele of scw1. We confirmed this identification by showing (i) that ng124 and an scw1 deletion fail to complement in a diploid (Figure 5C), (ii) that two plasmids containing only scw1+ in their region of overlap rescue the ng124 phenotype (Figure 5D), and (iii) that an ng124 strain has a G-to-A mutation (producing a stop codon) in codon 276 of the 562-codon scw1 ORF. Like ng124, scw1Δ has a strong synthetic effect with an spnΔ mutation (Table 3). Consistent with previous evidence (Karagiannis et al. 2002; Jin and McCollum 2003), our data suggest that Scw1 does not function in the AMR pathway: AMR formation and constriction had no obvious defects in an scw1Δ strain, and synthetic growth defects were seen between scw1-ng124 and both myo2-E1 and cdc4-s16 (data not shown). Taken together, our data suggest that Scw1 functions in parallel to the AMR and the septin/Mid2 pathway to promote cell division (see discussion).
DISCUSSION
The roles of the septins remain among the central puzzles of cytokinesis. These proteins localize to the division site in every fungal and animal cell type examined, and they are essential for successful cytokinesis in many cases. In other cell types, however, the septins are dispensable for cytokinesis (see Introduction). One possible resolution of this paradox is that a conserved septin role in cytokinesis can also be filled effectively in some cell types by a redundant protein(s) or pathway. In an attempt to identify such a redundant system, we conducted a genetic screen in S. pombe for mutations that are lethal in combination with a loss of septins. Although the results obtained have not yet solved the central problem, they have nonetheless shed considerable light on the role of the septins and other aspects of S. pombe cell division. In particular, the results suggest strongly that a septin/Mid2-dependent pathway functions in parallel with several other pathways to ensure successful cytokinesis, septation, and cell separation (Figure 7).
Possible organization of parallel pathways that cooperate to allow successful cytokinesis, septation, and cell separation in fission yeast, as suggested by the results of this study. Boldface type indicates the products of genes identified in this study. The cell-wall-integrity pathway headed by Wsc1 contains additional proteins beyond those shown here; the degree to which this pathway participates in unperturbed cytokinesis is not known. See text for details.
Genetic interaction between the septins and Cdc4:
One of the synthetic-lethal mutants contained a hypomorphic or neomorphic allele, cdc4-s16, of the essential gene cdc4. Cdc4 is thought to function as a light chain for both type II myosin heavy chains, Myo2 and Myp2 (Naqvi et al. 1999; Motegi et al. 2000; D'Souza et al. 2001; Lord and Pollard 2004). Myo2 and Myp2 appear to be partially redundant for function in cytokinesis but are specialized such that myo2 is an essential gene whereas myp2 is essential only under particular growth conditions (Bezanilla and Pollard 2000; Motegi et al. 2000). We found that cdc4-s16 and myp2Δ, like myo2-E1 and myp2Δ, are synthetically lethal (Figure 2, A–D); this suggests that cdc4-s16 compromises the function of Myo2, consistent with the other strong evidence that Cdc4 is a light chain for Myo2. We also found that cdc4-s16 and myo2-E1 are synthetically lethal (see results). Similar observations have been made with two other cdc4 alleles (Naqvi et al. 1999). Because myp2Δ is also synthetically lethal with myo2-E1, these results suggest that cdc4-s16 compromises the function of Myp2, consistent with the other evidence that Cdc4 is a light chain for Myp2. The hypothesis that cdc4-s16 generally compromises myosin II function in the AMR is also supported by the observation that cdc4-s16, like a variety of other mutations affecting the AMR (Mishra et al. 2004), is synthetically lethal with a clp1Δ protein phosphatase mutation (our unpublished results). However, other explanations for the cdc4-s16 myo2-E1 synthetic lethality remain possible. For example, it might result from an exacerbation by cdc4-s16 of the myo2-E1 effect on Myo2 function or from a partial loss of AMR function (due to myo2-E1) combined with a partial loss of some other Cdc4-mediated function (see below).
It is possible that the synthetic lethality between cdc4-s16 and septin mutations arises because the lack of septins further compromises the function of Myo2 or Myp2. However, we could obtain no evidence to support these possibilities (see results and Figure 2, E and F). Another hypothesis is that cdc4-s16 also compromises the function of some other Cdc4-binding partner that is part of a pathway that normally complements the pathway involving the septins. In this regard, it should be noted that there is good evidence that both S. pombe Cdc4 and its S. cerevisiae counterpart, Mlc1, interact with partners other than the type II myosin heavy chains and that these other partners are also important for cytokinesis (Stevens and Davis 1998; Boyne et al. 2000; Shannon and Li 2000; Desautels et al. 2001; D'Souza et al. 2001; Motegi et al. 2001; Win et al. 2001; Lord and Pollard 2004; Luo et al. 2004; Mulvihill et al. 2006; Park et al. 2009). The other partners include type V myosin heavy chains (S. pombe Myo51 and perhaps Myo52; S. cerevisiae Myo2), which are thought to be involved in the delivery of new plasma-membrane and cell-wall materials to the division site or (for Myo51) in some unknown function within the AMR; IQGAPs (S. pombe Rng2 and S. cerevisiae Iqg1), which may also be involved in plasma-membrane and cell-wall formation in addition to their roles in AMR function; and a phosphatidylinositol 4-kinase (S. pombe Pik1). From this hypothesis, it would be expected that the spnΔ cdc4-s16 synthetic lethality would be paralleled by strong synthetic effects between spnΔ and mutations affecting these other binding partners. However, we found no detectable synthetic effect between spn1-Δ2 and myo51Δ, rng2-D5 (Balasubramanian et al. 1998; Eng et al. 1998), rng2-346 (Chang et al. 1996), or pik1Δ117-198 (Onishi et al. 2010) and only a weak synthetic effect between spn1-Δ2 and myo52Δ (our unpublished results).
Thus, the most attractive hypothesis to explain the available data is that the septins are involved in a pathway whose function normally complements that of the AMR (Figure 7). If cdc4-s16 indeed compromises the function of both Myo2 and Myp2, its overall effect on the AMR may be sufficiently severe to produce synthetic lethality in combination with a loss of the septin-dependent pathway. However, given the lack of an obvious synthetic effect in a myo2-E1 myp2Δ spn1Δ triple mutant (Figure 2, E and F), this hypothesis also requires us to suppose that the cdc4-s16 effect on the AMR is more complex than a simple loss of myosin II function. Thus, it remains possible that the synthetic lethality between cdc4-s16 and septin mutations reflects an effect of cdc4-s16 on some unknown Cdc4 binding partner; further studies will be necessary to clarify this situation. In any case, however, the results do demonstrate the important point that the septins have a role in S. pombe cytokinesis that becomes critical under some conditions.
Role of a cell-wall-integrity pathway in cytokinesis and in the viability of septin mutants:
Five of the remaining synthetic-lethal mutants had very similar cell-lysis phenotypes and mapped to two loci. Our attempts to clone the corresponding genes led to the isolation of numerous dosage suppressors (Table 4), in addition to the gene (rgf3) that actually contains the s44 mutation. Given what is known about the functions of Rho1, the Rho-GEFs, Pck2, and Pob1 in S. pombe (see Introduction; Toya et al. 1999; Rincón et al. 2009) and about the well-studied cell-wall-integrity pathway in S. cerevisiae (Levin 2005), it seems clear that the seven genes identified as dosage suppressors all represent components of a similar pathway in S. pombe, as also described, in part, in several other recent studies (Madrid et al. 2006, 2007; Barba et al. 2008; and references cited therein). In this model, Wsc1 is a sensor of cell-wall damage that works through one or more of the Rho-GEFs Rgf1, Rgf2, and Rgf3 to activate Rho1. (Note that a Rho-GEF that does not normally function in this pathway might be able to do so when overexpressed.) Activated Rho1 (with its function facilitated by Pob1) would trigger several responses that would help to correct the damage, including the activation of Pck2, which would in turn activate a MAP-kinase pathway (Madrid et al. 2006; Barba et al. 2008) that would lead to increased expression of genes whose products are involved in the repair of cell-wall damage.
The identification of s34 as a mutation in SPBC19G7.08c/art1, which encodes one of a family of at least eight arrestin-related proteins in S. pombe, was unexpected and interesting. The arrestins were originally identified for their roles in the regulation of G-protein-coupled receptors but were later found to function more broadly in the endocytic regulation of plasma-membrane proteins (Lefkowitz et al. 2006; Moore et al. 2007). To our knowledge, the S. pombe arrestin-related proteins have not been studied previously, except for one involved in regulating meiotic entry (Matsuyama et al. 2000), but their counterparts in S. cerevisiae have been reported to function as ubiquitin-ligase adaptors that regulate endocytosis of cell-surface proteins (Lin et al. 2008; Nikko and Pelham 2009). The observations that Art1 localizes to the cell-division site, that the art1-s34 mutant phenotype is essentially identical to that of rgf3-s44, and that art1-s34 is rescued by overexpression of seven different genes encoding components of the cell-wall-integrity pathway all suggest strongly that Art1 also functions as a component of this pathway. Thus, our continuing studies of Art1 function should help to elucidate not only the mechanisms for ensuring cell-wall integrity during cell division but also the functions of this family of proteins.
In any case, a plausible interpretation of our observations to date is as follows (Figure 7). In the absence of the septins, the septum that forms has defects, the localization of lytic enzymes that function in cell separation is perturbed (see Martín-Cuadrado et al. 2005; Sipiczki 2007), or both. If the cell-wall-integrity pathway is functioning normally, the problems can be recognized and repaired, and the cells usually divide successfully (albeit typically after a significant delay). The rgf3-s44 and art1-s34 mutations compromise the cell-wall-integrity pathway, so that the damage resulting from the absence of septins is not repaired efficiently, and thus the cells often lyse when they attempt to separate. However, overexpression of components of the cell-wall-integrity pathway can activate it sufficiently that the absence of other components is overcome, cell-wall problems are repaired, and cell division usually occurs successfully. Given that the MAP-kinase pathway appears to be partially activated in every cell cycle (Madrid et al. 2007), the absence of the septins may only exacerbate a problem that exists even in normal cells as they attempt the delicate task of forming a septum and then degrading it enough to separate, without leaving a weakness that would lead to lysis driven by the intracellular turgor pressure.
In this context, it is also of interest that, in the scw1-ng124 mutant, some cells underwent seemingly normal cell separation long after the septins had disassembled from the septal region (Figure 5B, cell 1 at 00:00, 01:17, and 2:33). This observation suggests that the septins may not play a direct role in cell separation and that some checkpoint mechanism delays attempted cell separation in an scw1-ng124 (or spnΔ) mutant until septal-wall defects have been repaired.
The possibility of a pathway redundant with that involving the septins:
We began the synthetic-lethal screen with the hope of identifying a functionally redundant pathway that would explain why the septins are not essential for cytokinesis in S. pombe and in some other cell types. It is possible that the final mutation identified in our screen, scw1-ng124, affects such a pathway (Figure 7). The scw1-ng124 and scw1Δ single mutants have a division delay like that seen in septin or mid2 mutants, but several observations suggest that Scw1 does not function in the septin/Mid2 pathway. First, although the septins and Mid2 depend on each other for normal localization, both the septins and Mid2 localize essentially normally in an scw1 mutant (Figure 6). Moreover, although septin and mid2 mutations have no synthetic effect with each other, scw1 mutations have strong synthetic effects with each (Figures 5A and 6; Table 3).
Scw1 and scw1 mutants have been described previously (Karagiannis et al. 2002; Jin and McCollum 2003). Scw1 contains a putative RNA-binding domain, and an S. cerevisiae homolog, Whi3, indeed binds to a large set of mRNAs (Colomina et al. 2008). Because scw1 mutations can suppress mutations both in the SIN pathway (see Introduction) and in the β-glucan synthase Bgs1, the earlier reports suggested that Scw1 might be a negative regulator of septum formation, so that scw1 mutations would promote septum formation. However, this interpretation is difficult to reconcile with the observations that the septa in an scw1 mutant seem to appear with normal timing but then be delayed in separation (Figure 5B) and that this cell-separation delay is enhanced in scw1 spn and scw1 mid2 double mutants (Figures 1P, 5A, and 6I; Table 3). Thus, we suggest that a more likely interpretation of the available information is as follows. Scw1 may promote some step in the initiation of cell separation or a step in septum completion (perhaps not easy to visualize microscopically) that is necessary to initiate cell separation. Thus, the absence of functional Scw1 in an scw1 mutant would (1) result in multiseptated cell chains, as observed, and (2) rescue SIN mutants and bgs1-191 by producing a delay in attempted cell separation that would provide more time for SIN- and Bgs1-dependent processes to be completed, so that the daughter cells would separate successfully without lysis. If Scw1 and the septins function in parallel pathways for completion of the same events, then the strong synthetic effect between scw1Δ and spnΔ (or mid2Δ) mutations could be explained. The identification of proteins and/or mRNAs bound by Scw1 should enable us to explore these possibilities further.
Limitations of the synthetic-lethal screening to date:
Because we obtained only one cdc4 and one scw1 mutation, our screen was probably not saturated, so that further screening of the same type might identify additional genes of interest. However, it is also possible that the design of the screen limited the genes that we were able to identify. Use of the nmt1 promoter for synthetic-lethal screening is convenient in the simple comparison of cells grown in the absence and presence of thiamine (Yoon et al. 1997; Berry et al. 1999; this study). However, the nmt1 promoters are leaky, and cells retain some level of expression under repressing conditions (Forsburg 1993; Berry et al. 1999). Thus, we might have missed some mutations that would have had stronger interactions with septin null mutations. Further synthetic-lethal screening using a different system might be fruitful in providing additional insights into the roles of the septins in cytokinesis in S. pombe and thus, presumably, also in other types of cells.
Footnotes
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.119842/DC1.
Footnotes
Communicating editor: M. D. Rose
Acknowledgements
We thank Damian Brunner, Paul Nurse, Jianhua Liu, Mohan Balasubramanian, Susan Forsburg, Won-Ki Huh, Erin K. O'Shea, Roger Tsien, Paul Young, Antony Carr, Quan-wen Jin, Dan McCollum, Ken Sawin, Val Wood, and the Yeast Genetic Resource Center Japan (supported by the National BioResource Project) for strains, plasmids, libraries, and/or the communication of unpublished results; Chloé Diamond and Masayuki Onishi for help with strain constructions and for evaluating a mutant phenotype; Carlo Caruso for technical assistance; and members of our laboratories for helpful discussions. We also thank two anonymous reviewers of earlier versions of this article for insightful and helpful suggestions. This work was supported by National Institutes of Health grants GM-31006 to J.R.P., GM-26132 to T.D.P., and GM-086546 to J.-Q.W. and by a Basil O'Connor Starter Scholar Research Award to J.-Q.W.
References
Adam, J. C., J. R. Pringle and M. Peifer,
An, H., J. L. Morrell, J. L. Jennings, A. J. Link and K. L. Gould,
Anders, A., S. Watt, J. Bähler and K. E. Sawin,
Arellano, M., A. Durán and P. Pérez,
Arellano, M., A. Durán and P. Pérez,
Arellano, M., M. H. Valdivieso, T. M. Calonge, P. M. Coll, A. Durán et al.,
Bähler, J., J.-Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, III et al.,
Balasubramanian, M. K., D. McCollum and K. L. Gould,
Balasubramanian, M. K., D. McCollum, L. Chang, K. C. Wong, N. I. Naqvi et al.,
Balasubramanian, M. K., E. Bi and M. Glotzer,
Barba, G., T. Soto, M. Madrid, A. Núñez, J. Vicente et al.,
Basi, G., E. Schmid and K. Maundrell,
Berlin, A., A. Paoletti and F. Chang,
Berry, L. D., A. Feoktistova, M. D. Wright and K. L. Gould,
Bertin, A., M. A. McMurray, P. Grob, S.-S. Park, G. García, III et al.,
Bezanilla, M., and T. D. Pollard,
Bezanilla, M., S. L. Forsburg and T. D. Pollard,
Bezanilla, M., J. M. Wilson and T. D. Pollard,
Bi, E., P. Maddox, D. J. Lew, E. D. Salmon, J. N. McMillan et al.,
Boyne, J. R., H. M. Yosuf, P. Bieganowski, C. Brenner and C. Price,
Calonge, T. M., K. Nakano, M. Arellano, R. Arai, S. Katayama et al.,
Calonge, T. M., M. Arellano, P. M. Coll and P. Pérez,
Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird et al.,
Caudron, F., and Y. Barral,
Chang, F., A. Wollard and P. Nurse,
Colomina, N., F. Ferrezuelo, H. Wang, M. Aldea and E. Garí,
Cortés, J. C. G., E. Carnero, J. Ishiguro, Y. Sánchez, A. Durán et al.,
Desautels, M., J. P. Den Haese, C. M. Slupsky, L. P. McIntosh and S. M. Hemmingsen,
Dobbelaere, J., and Y. Barral,
D'Souza, V. M., N. I. Naqvi, H. Wang and M. K. Balasubramanian,
Eng, K., N. I. Naqvi, K. C. Wong and M. K. Balasubramanian,
Field, C. M., A. S. Maddox, J. R. Pringle and K. Oegema,
Forsburg, S. L.,
García, P., V. Tajadura, I. García and Y. Sánchez,
García, P., I. García, F. Marcos, G. R. de Garibay and Y. Sánchez,
García, P., V. Tajadura and Y. Sanchez,
Giansanti, M. G., R. M. Farkas, S. Bonaccorsi, D. L. Lindsley, B. T. Wakimoto et al.,
Gladfelter, A. S., J. R. Pringle and D. J. Lew,
Gould, K. L., and V. Simanis,
Hartwell, L. H.,
Hirata, D., K. Nakano, M. Fukui, H. Takenaka, T. Miyakawa et al.,
Hochstenbach, F., F. M. Klis, H. van den Ende, E. van Donselaar, P. J. Peters et al.,
Huh, W.-K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al.,
Humbel, B. M., M. Konomi, T. Takagi, N. Kamasawa, S. A. Ishijima et al.,
Ishiguro, J., A. Saitou, A. Durán and J. C. Ribas,
Iwaki, N., K. Karatsu and M. Miyamoto,
Jin, Q.-W., and D. McCollum,
John, C. M., R. K. Hite, C. S. Weirich, D. J. Fitzgerald, H. Jawhari et al.,
Karagiannis, J., R. Oulton and P. G. Young,
Katayama, S., D. Hirata, M. Arellano, P. Pérez and T. Toda,
Kinoshita, M., S. Kumar, A. Mizoguchi, C. Ide, A. Kinoshita et al.,
Kitayama, C., A. Sugimoto and M. Yamamoto,
Korinek, W. S., E. Bi, J. A. Epp, L. Wang, J. Ho et al.,
Krapp, A., and V. Simanis,
Lefkowitz, R. J., K. Rajagopal and E. J. Whalen,
Le Goff, X., A. Woollard and V. Simanis,
Leupold, U.,
Levin, D. E.,
Lin, C. H., J. A. MacGurn, T. Chu, C. J. Stefan and S. D. Emr,
Lippincott, J., and R. Li,
Lippincott, J., K. B. Shannon, W. Shou, R. J. Deshaies and R. Li,
Liu, J., H. Wang, D. McCollum and M. K. Balasubramanian,
Liu, J., H. Wang and M. K. Balasubramanian,
Liu, J., X. Tang, H. Wang, S. Oliferenko and M. K. Balasubramanian,
Longtine, M. S., D. J. DeMarini, M. L. Valencik, O. S. Al-Awar, H. Fares et al.,
Lord, M., and T. D. Pollard,
Luo, J., E. A. Vallen, C. Dravis, S. E. Tcheperegine, B. Drees et al.,
Ma, Y., T. Kuno, A. Kita, Y. Asayama and R. Sugiura,
Maddox, A. S., L. Lewellyn, A. Desai and K. Oegema,
Maddox, P. S., K. S. Bloom and E. D. Salmon,
Madrid, M., T. Soto, H. K. Khong, A. Franco, J. Vicente et al.,
Madrid, M., A. Núñez, T. Soto, J. Vicente-Soler, M. Gacto et al.,
Martín, V., B. García, E. Carnero, A. Durán and Y. Sánchez,
Martín-Cuadrado, A. B., J. L. Morrell, M. Konomi, H. An, C. Petit et al.,
Matsuyama, A., N. Yabana, Y. Watanabe and M. Yamamoto,
McCollum, D., and K. L. Gould,
McCollum, D., M. K. Balasubramanian, L. E. Pelcher, S. M. Hemmingsen and K. L. Gould,
McMurray, M. A., and J. Thorner,
Miki, T., C. L. Smith, J. E. Long, A. Eva and T. P. Fleming,
Mishra, M., J. Karagiannis, S. Trautmann, H. Wang, D. McCollum et al.,
Moore, C. A., S. K. Milano and J. L. Benovic,
Moreno, S., A. Klar and P. Nurse,
Morrell-Falvey, J. L., L. Ren, A. Feoktistova, G. D. Haese and K. L. Gould,
Motegi, F., K. Nakano and I. Mabuchi,
Motegi, F., R. Arai and I. Mabuchi,
Mulvihill, D. P., S. R. Edwards and J. S. Hyams,
Mutoh, T., K. Nakano and I. Mabuchi,
Nakano, K., R. Arai and I. Mabuchi,
Nakano, K., T. Mutoh and I. Mabuchi,
Naqvi, N. I., K. Eng, K. L. Gould and M. K. Balasubramanian,
Neufeld, T. P., and G. M. Rubin,
Nguyen, T. Q., H. Sawa, H. Okano and J. G. White,
Nikko, E., and H. R. B. Pelham,
Nurse, P., P. Thuriaux and K. Nasmyth,
O'Keefe, L., W. G. Somers, A. Harley and R. Saint,
Onishi, M., T. Koga, A. Hirata, T. Nakamura, H. Asakawa et al.,
Pan, F., R. L. Malmberg and M. Momany,
Park, J. S., S. K. Steinbach, M. Desautels and S. M. Hemmingsen,
Petit, C. S., S. Mehta, R. H. Roberts and K. L. Gould,
Pollard, T. D., and J.-Q. Wu,
Prokopenko, S. N., A. Brumby, L. O'Keefe, L. Prior, Y. He et al.,
Rincón, S. A., Y. Ye, M. A. Villar-Tajadura, B. Santos, S. G. Martin et al.,
Sayers, L. G., S. Katayama, K. Nakano, H. Mellor, I. Mabuchi et al.,
Schmidt, H.,
Schmidt, M., B. Bowers, A. Varma, D. H. Roh and E. Cabib,
Shandala, T., S. L. Gregory, H. E. Dalton, M. Smallhorn and R. Saint,
Shannon, K. B., and R. Li,
Sipiczki, M.,
Sirajuddin, M., M. Farkasovsky, F. Hauer, D. Kühlmann, I. G. Macara et al.,
Stevens, R. C., and T. N. Davis,
Surka, M. C., C. W. Tsang and W. S. Trimble,
Tajadura, V., B. García, I. García, P. García and Y. Sánchez,
Tasto, J. J., J. L. Morrell and K. L. Gould,
Tatsumoto, T., X. Xie, R. Blumenthal, I. Okamoto and T. Miki,
Toya, M., Y. Iino and M. Yamamoto,
Win, T. Z., Y. Gachet, D. P. Mulvihill, K. M. May and J. S. Hyams,
Wright, A., K. Maundrell, W. D. Heyer, D. Beach and P. Nurse,
Wu, J.-Q., J. Bähler and J. R. Pringle,
Wu, J.-Q., J. R. Kuhn, D. R. Kovar and T. D. Pollard,
Yang, F., L. G. Moss and G. N. Phillips, Jr.,
Yoon, J. H., W. A. Whalen, A. Bharathi, R. Shen and R. Dhar,
Zacharias, D. A., J. D. Violin, A. C. Newton and R. Y. Tsien,