-
PDF
- Split View
-
Views
-
Cite
Cite
Christine L Kerwin, Dennis D Wykoff, Candida glabrata PHO4 Is Necessary and Sufficient for Pho2-Independent Transcription of Phosphate Starvation Genes, Genetics, Volume 182, Issue 2, 1 June 2009, Pages 471–479, https://doi.org/10.1534/genetics.109.101063
Close - Share Icon Share
Abstract
Comparative genomic analyses of Candida glabrata and Saccharomyces cerevisiae suggest many signal transduction pathways are highly conserved. Focusing on the phosphate signal transduction (PHO) pathway of C. glabrata, we demonstrate that components of the pathway are conserved and confirm the role of CgPHO81, CgPHO80, CgPHO4, and CgMSN5 in the PHO pathway through deletion analysis. Unlike S. cerevisiae, C. glabrata shows little dependence on the transcription factor, Pho2, for induction of phosphate-regulated genes during phosphate limitation. We show that the CgPho4 protein is necessary and sufficient for Pho2-independent gene expression; CgPho4 is capable of driving expression of PHO promoters in S. cerevisiae in the absence of ScPHO2. On the basis of the sequences of PHO4 in the hemiascomycetes and complementation analysis, we suggest that Pho2 dependence is a trait only observed in species closely related to S. cerevisiae. Our data are consistent with trans-regulatory changes in the PHO pathway via the transcription factor Pho4 as opposed to cis-regulatory changes (the promoter).
DIFFERENT species exploit specific niches. Gene expression programs have evolved to allow for optimal growth in these niches. Many gene expression programs are common to species (e.g., DNA damage response, or nutrient starvation response); however, it is unclear how signal transduction pathways have evolved to tailor gene expression programs to their niche (Fry et al. 2006; Gasch 2007). Traditional signal transduction studies utilize conservation of signaling components in one species to infer conservation of functional output and this has been successful at predicting behaviors of important signal transduction pathways in organisms ranging from humans to bacteria (Simon 2001; Pinter et al. 2005). However, recent work has demonstrated that even when components are highly conserved between organisms, the network architecture, or how the components may interact, is different. For example, chemotaxis in Escherichia coli and Bacillus subtilis relies on the same components but the interactions are different, resulting in divergent behavior (Alon et al. 1999; Rao et al. 2004). This is underscored by examples of signal transduction pathways that behave differently between humans and mice (Migeon et al. 2005; Garofalo 2006). Thus, comparative analyses of signaling pathways are the first step in establishing the evolutionary pressures regulating speciation. Multicellular systems (notably the drosophilids) present a diversity of sequenced organisms and molecular genetic tools; however, the complexity of pathways, the number of components, and the exponential number of possible network interactions makes a thorough comparative analysis difficult (Yeger-Lotem et al. 2004; Moses et al. 2006).
The complete genomic sequence of many ascomycetes has begun to allow for a comparative genomic approach to understand the evolutionary steps required for speciation (Cliften et al. 2003; Dujon et al. 2004). For example, if a species does not experience an environmental condition during evolutionary time, the pathway required to respond to that condition can decay (Hittinger et al. 2004). Furthermore, promoters responsive to important signaling pathways, such as ribosomal protein biogenesis, can drift and acquire the ability to bind different transcription factors (Tsong et al. 2003; Butler et al. 2004; Ihmels et al. 2005). In this study, we have utilized a comparative genomic approach to study the PHO pathway in Candida glabrata and contrast its signaling pathway with the well studied PHO pathway of Saccharomyces cerevisiae.
The PHO pathway in S. cerevisiae activates the transcription of at least 20 genes during phosphate starvation (Lenburg and O'Shea 1996; Oshima 1997; Carroll et al. 2001). The PHO pathway consists of upstream signaling components (Pho81, Pho80, Pho85, Pho4, and Pho2) and a downstream transcriptional output. For the purposes of this study, we examine the transcription of PHO5 and PHO84 in S. cerevisiae and PHO84 and GIT1 in C. glabrata. Core to the signaling pathway is Pho4, a transcription factor regulated by a cyclin/cyclin-dependent kinase (CDK)/CDK inhibitor complex composed of Pho80, Pho85, and Pho81. In high phosphate conditions, the Pho80/Pho85 complex phosphorylates Pho4 on four key serine residues, causing export of Pho4 by Msn5 from the nucleus and cytoplasmic localization (O'Neill et al. 1996; Komeili and O'Shea 1999). In low phosphate conditions, Pho81 inhibits the kinase complex causing dephosphorylated Pho4 to accumulate in the nucleus via the import receptor Pse1, allowing Pho4 to cooperatively bind phosphate starvation promoters with the transcriptional coactivator Pho2 (Vogel et al. 1989; Yoshida et al. 1989a; Schneider et al. 1994).
We examined the PHO pathway in C. glabrata, because it is closely related to S. cerevisiae in the ascomycete lineage; however, both species experienced an ancestral whole genome duplication supplying the raw material for neofunctionalization (Wolfe 2001; Dujon et al. 2004). Because these species diverged from a common ancestor ∼10 MYA, share ∼75% protein sequence identity, and share many signal transduction pathways, we hypothesized C. glabrata would have many similarities to S. cerevisiae, with the notable difference of environment; C. glabrata is a commensal pathogen with mammals (Redondo-Lopez et al. 1990; Cormack and Falkow 1999; Cormack et al. 1999; Domergue et al. 2005).
We demonstrate that whereas most components of the PHO pathway are conserved between the two species, an important difference exists that may affect speciation. Pho2 is not as important for the transcriptional response to phosphate starvation in C. glabrata as it is in S. cerevisiae. We propose this Pho2 requirement (or lack of) has evolved in trans and that mutations in promoters have had a minimal impact on the Pho2 requirement as is evidenced by the sufficiency of CgPho4 to circumvent the Pho2 requirement in S. cerevisiae. (We abbreviate the derivation of genes from the two organisms as Sc, S. cerevisiae and Cg, C. glabrata.) Comparative genomic analysis of the hemiascomycetes suggests that the requirement for Pho2 in the transcriptional induction of PHO genes in S. cerevisiae is a derived trait, and the inclusion of Pho2 into the PHO pathway could have allowed for the Saccharomyces genus to take advantage of low inorganic phosphate conditions in nature.
MATERIALS AND METHODS
Strain construction:
C. glabrata mutants were generated using antibiotic resistance genes KANMX6 or NATMX6 and homologous recombination to inactivate various phosphate signaling genes in a C. glabrata his3− background (Longtine et al. 1998; Cormack and Falkow 1999; Hentges et al. 2005). See Table 1 for a summary of strains and supporting information, Table S1, for a description of primers utilized to generate strains and plasmids. Deletion of genes was confirmed by PCR. The mutant strains were further confirmed by a semiquantitative phosphatase assay to confirm that multiple isolates behaved similarly.
Strains used in this study
Strain . | Genotype . | Reference . |
|---|---|---|
| S. cerevisiae | ||
| EY57 | K699 with MATa | Wykoff and O'Shea (2001) |
| EY131 | EY57 with pho4ΔTRP1 | Wykoff et al. (2007) |
| EY337 | EY57 with pho2ΔLEU2 | Wykoff et al. (2007) |
| EY338 | EY57 with pho4ΔTRP1 pho2ΔLEU2 | Wykoff et al. (2007) |
| C. glabrata | ||
| BG99 | his3Δ(1 + 631) | Cormack and Falkow (1999) |
| DG2 | BG99 with Cgpho4ΔKANMX6 | This study |
| DG3 | BG99 with Cgpho2ΔKANMX6 | This study |
| DG6 | BG99 with Cgpho81ΔKANMX6 | This study |
| DG8 | BG99 with Cgpho80ΔNATMX6 | This study |
| DG9 | BG99 with Cgmsn5ΔNATMX6 | This study |
| DG11 | BG99 with Cgpho4ΔKANMX6 Cgpho2ΔNATMX6 | This study |
Strain . | Genotype . | Reference . |
|---|---|---|
| S. cerevisiae | ||
| EY57 | K699 with MATa | Wykoff and O'Shea (2001) |
| EY131 | EY57 with pho4ΔTRP1 | Wykoff et al. (2007) |
| EY337 | EY57 with pho2ΔLEU2 | Wykoff et al. (2007) |
| EY338 | EY57 with pho4ΔTRP1 pho2ΔLEU2 | Wykoff et al. (2007) |
| C. glabrata | ||
| BG99 | his3Δ(1 + 631) | Cormack and Falkow (1999) |
| DG2 | BG99 with Cgpho4ΔKANMX6 | This study |
| DG3 | BG99 with Cgpho2ΔKANMX6 | This study |
| DG6 | BG99 with Cgpho81ΔKANMX6 | This study |
| DG8 | BG99 with Cgpho80ΔNATMX6 | This study |
| DG9 | BG99 with Cgmsn5ΔNATMX6 | This study |
| DG11 | BG99 with Cgpho4ΔKANMX6 Cgpho2ΔNATMX6 | This study |
The genotype of K699 is ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3.
Strains used in this study
Strain . | Genotype . | Reference . |
|---|---|---|
| S. cerevisiae | ||
| EY57 | K699 with MATa | Wykoff and O'Shea (2001) |
| EY131 | EY57 with pho4ΔTRP1 | Wykoff et al. (2007) |
| EY337 | EY57 with pho2ΔLEU2 | Wykoff et al. (2007) |
| EY338 | EY57 with pho4ΔTRP1 pho2ΔLEU2 | Wykoff et al. (2007) |
| C. glabrata | ||
| BG99 | his3Δ(1 + 631) | Cormack and Falkow (1999) |
| DG2 | BG99 with Cgpho4ΔKANMX6 | This study |
| DG3 | BG99 with Cgpho2ΔKANMX6 | This study |
| DG6 | BG99 with Cgpho81ΔKANMX6 | This study |
| DG8 | BG99 with Cgpho80ΔNATMX6 | This study |
| DG9 | BG99 with Cgmsn5ΔNATMX6 | This study |
| DG11 | BG99 with Cgpho4ΔKANMX6 Cgpho2ΔNATMX6 | This study |
Strain . | Genotype . | Reference . |
|---|---|---|
| S. cerevisiae | ||
| EY57 | K699 with MATa | Wykoff and O'Shea (2001) |
| EY131 | EY57 with pho4ΔTRP1 | Wykoff et al. (2007) |
| EY337 | EY57 with pho2ΔLEU2 | Wykoff et al. (2007) |
| EY338 | EY57 with pho4ΔTRP1 pho2ΔLEU2 | Wykoff et al. (2007) |
| C. glabrata | ||
| BG99 | his3Δ(1 + 631) | Cormack and Falkow (1999) |
| DG2 | BG99 with Cgpho4ΔKANMX6 | This study |
| DG3 | BG99 with Cgpho2ΔKANMX6 | This study |
| DG6 | BG99 with Cgpho81ΔKANMX6 | This study |
| DG8 | BG99 with Cgpho80ΔNATMX6 | This study |
| DG9 | BG99 with Cgmsn5ΔNATMX6 | This study |
| DG11 | BG99 with Cgpho4ΔKANMX6 Cgpho2ΔNATMX6 | This study |
The genotype of K699 is ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3.
To generate PHO4 and PHO2 containing plasmids, the ORF for the gene with at least 500 bp upstream was PCR amplified with primers that created a NotI restriction site at the 5′ end and a PacI restriction site at the 3′ end of the gene. Primers used in this study are described in Table S1. The PCR fragment was digested with NotI and PacI and ligated in frame into pRS313-13myc, so that the myc epitope was C terminal (Sikorski and Hieter 1989; Wykoff and O'Shea 2005). For Figure 6, all PHO4 genes were amplified with a stop codon and were not in frame with the myc epitope. The PHO4 and PHO2 plasmids, for both S. cerevisiae and C. glabrata, were transformed into yeast strains using a standard lithium acetate yeast transformation protocol with selection on medium lacking histidine (Wykoff and O'Shea 2001; Guthrie and Fink 2002).
Media and growth conditions:
Yeast strains defective in PHO pathway genes, but without plasmids, were grown in synthetic dextrose (SD) media with complete supplement mixture (CSM) amino acids (Sunrise Science Products, San Diego, CA) at 30° until logarithmic growth phase (see Guthrie and Fink 2002 for media components). Logarithmic growth is an OD600 ∼0.5. Yeast strains defective in pathway genes and containing HIS3+ plasmids were grown in SD +CSM −histidine. For all described experiments, cells were grown to logrithmic phase, pelleted by centrifugation, washed three times in medium lacking phosphate, then transferred to media lacking phosphate or media with 10 mm KH2PO4 (for phosphate replete conditions) and grown at 30° for 3 hr.
Comparative genomic analysis:
Orthologs of each S. cerevisiae PHO pathway component were identified in C. glabrata on the basis of sequence similarity using the “Blastp vs. Fungi” feature in the Saccharomyces Genome Database (http://www.yeastgenome.org/). The result with the lowest expect (E) value (specifically a cutoff of E = 10−2) was determined to be the ortholog in C. glabrata. To obtain values useful in comparing the two species, the sequences for the orthologs in the two species were aligned using the National Center for Biotechnology Information (NCBI) Blastp and the BLOSUM62 matrix (http://www.ncbi.nlm.nih.gov/). The relevant values were recorded; specifically, percentage of amino acid identity, the number of amino acids over which the percentage of amino acid identity was measured, expect value, as well as the sizes of the proteins in S. cerevisiae and C. glabrata (Table 2).
Genomic sequence predicts a C. glabrata PHO pathway very similar to S. cerevisiae
. | . | Sc to Cg . | Length of protein . | ||
|---|---|---|---|---|---|
| Gene . | Cg systematic name . | Identity (%) . | Expect . | Sc . | Cg . |
| MSN5 | CAGL0M01144g | 935/1219 (76) | 0 | 1225 | 1221 |
| PHO84 | CAGL0B02475g | 448/586 (76) | 0 | 588 | 580 |
| PHO87 | CAGL0F02387g | 649/966 (67) | 0 | 924 | 952 |
| PHO90 | CAGL0F02387g | 624/957 (65) | 0 | 882 | 952 |
| PSE1 | CAGL0M13871g | 754/1091 (69) | 0 | 1090 | 1091 |
| VIP1 | CAGL0M09823g | 832/1113 (74) | 0 | 1147 | 1128 |
| PHO85 | CAGL0L12474g | 273/299 (91) | 3E-159 | 306 | 302 |
| PHO81 | CAGL0L06622g | 344/900 (38) | 5E-144 | 1179 | 1137 |
| ADK1 | CAGL0K11418g | 201/222 (90) | 6E-113 | 223 | 222 |
| PHO23 | CAGL0G06556g | 197/336 (58) | 5E-88 | 331 | 316 |
| PHO86 | CAGL0L05456g | 163/300 (54) | 5E-86 | 312 | 306 |
| PHO2 | CAGL0L07436g | 180/384 (46) | 8E-85 | 560 | 515 |
| PHO80 | CAGL0E02541g | 138/261 (52) | 7E-73 | 294 | 342 |
| PHO88 | CAGL0K12276g | 134/196 (68) | 8E-68 | 189 | 194 |
| PHO4 | CAGL0D05170g | 32/60 (53) | 4E-08 | 313 | 533 |
| SPL2 | No ortholog | ||||
| PHO89 | No ortholog | ||||
| NCP1 | CAGL0D04114g | 457/693 (65) | 0 | 692 | 687 |
| PHM1 | CAGL0F02145g | 499/831 (60) | 0 | 829 | 816 |
| PHM2 | CAGL0F02145g | 489/854 (57) | 0 | 836 | 816 |
| PHM3 | CAGL0G06952g | 582/719 (80) | 0 | 722 | 717 |
| PHO8 | CAGL0H07359g | 322/488 (65) | 4E-176 | 567 | 545 |
| AUT4 | CAGL0B00770g | 311/538 (57) | 1E-162 | 529 | 541 |
| PHO91 | CAGL0I05632g | 289/690 (41) | 8E-133 | 895 | 886 |
| SDT1 | CAGL0H09218g | 158/265 (59) | 6E-90 | 281 | 279 |
| IRC15 | CAGL0F01947g | 183/471 (38) | 5E-79 | 500 | 493 |
| GIT1 | CAGL0A01243g | 146/483 (30) | 3E-60 | 519 | 531 |
| PHM8 | CAGL0C02321g | 136/292 (46) | 2E-58 | 322 | 314 |
| PHM4 | CAGL0M12705g | 110/124 (88) | 1E-53 | 130 | 127 |
| CTF19 | CAGL0F02035g | 105/334 (31) | 6E-32 | 370 | 373 |
| PHM6 | No ortholog | ||||
| PHO5 | No ortholog | ||||
| PHO11 | No ortholog | ||||
| PHO12 | No ortholog | ||||
. | . | Sc to Cg . | Length of protein . | ||
|---|---|---|---|---|---|
| Gene . | Cg systematic name . | Identity (%) . | Expect . | Sc . | Cg . |
| MSN5 | CAGL0M01144g | 935/1219 (76) | 0 | 1225 | 1221 |
| PHO84 | CAGL0B02475g | 448/586 (76) | 0 | 588 | 580 |
| PHO87 | CAGL0F02387g | 649/966 (67) | 0 | 924 | 952 |
| PHO90 | CAGL0F02387g | 624/957 (65) | 0 | 882 | 952 |
| PSE1 | CAGL0M13871g | 754/1091 (69) | 0 | 1090 | 1091 |
| VIP1 | CAGL0M09823g | 832/1113 (74) | 0 | 1147 | 1128 |
| PHO85 | CAGL0L12474g | 273/299 (91) | 3E-159 | 306 | 302 |
| PHO81 | CAGL0L06622g | 344/900 (38) | 5E-144 | 1179 | 1137 |
| ADK1 | CAGL0K11418g | 201/222 (90) | 6E-113 | 223 | 222 |
| PHO23 | CAGL0G06556g | 197/336 (58) | 5E-88 | 331 | 316 |
| PHO86 | CAGL0L05456g | 163/300 (54) | 5E-86 | 312 | 306 |
| PHO2 | CAGL0L07436g | 180/384 (46) | 8E-85 | 560 | 515 |
| PHO80 | CAGL0E02541g | 138/261 (52) | 7E-73 | 294 | 342 |
| PHO88 | CAGL0K12276g | 134/196 (68) | 8E-68 | 189 | 194 |
| PHO4 | CAGL0D05170g | 32/60 (53) | 4E-08 | 313 | 533 |
| SPL2 | No ortholog | ||||
| PHO89 | No ortholog | ||||
| NCP1 | CAGL0D04114g | 457/693 (65) | 0 | 692 | 687 |
| PHM1 | CAGL0F02145g | 499/831 (60) | 0 | 829 | 816 |
| PHM2 | CAGL0F02145g | 489/854 (57) | 0 | 836 | 816 |
| PHM3 | CAGL0G06952g | 582/719 (80) | 0 | 722 | 717 |
| PHO8 | CAGL0H07359g | 322/488 (65) | 4E-176 | 567 | 545 |
| AUT4 | CAGL0B00770g | 311/538 (57) | 1E-162 | 529 | 541 |
| PHO91 | CAGL0I05632g | 289/690 (41) | 8E-133 | 895 | 886 |
| SDT1 | CAGL0H09218g | 158/265 (59) | 6E-90 | 281 | 279 |
| IRC15 | CAGL0F01947g | 183/471 (38) | 5E-79 | 500 | 493 |
| GIT1 | CAGL0A01243g | 146/483 (30) | 3E-60 | 519 | 531 |
| PHM8 | CAGL0C02321g | 136/292 (46) | 2E-58 | 322 | 314 |
| PHM4 | CAGL0M12705g | 110/124 (88) | 1E-53 | 130 | 127 |
| CTF19 | CAGL0F02035g | 105/334 (31) | 6E-32 | 370 | 373 |
| PHM6 | No ortholog | ||||
| PHO5 | No ortholog | ||||
| PHO11 | No ortholog | ||||
| PHO12 | No ortholog | ||||
Orthologs of the PHO pathway components, both regulators and transcriptional output, in S. cerevisiae (Sc) were determined for C. glabrata (Cg) on the basis of sequence similarity. Components are listed from lowest expect value to highest expect value and components with no ortholog are listed last. MSN5 through PHO89 are signaling components and NCP1 through PHO12 are genes upregulated in response to phosphate starvation.
Genomic sequence predicts a C. glabrata PHO pathway very similar to S. cerevisiae
. | . | Sc to Cg . | Length of protein . | ||
|---|---|---|---|---|---|
| Gene . | Cg systematic name . | Identity (%) . | Expect . | Sc . | Cg . |
| MSN5 | CAGL0M01144g | 935/1219 (76) | 0 | 1225 | 1221 |
| PHO84 | CAGL0B02475g | 448/586 (76) | 0 | 588 | 580 |
| PHO87 | CAGL0F02387g | 649/966 (67) | 0 | 924 | 952 |
| PHO90 | CAGL0F02387g | 624/957 (65) | 0 | 882 | 952 |
| PSE1 | CAGL0M13871g | 754/1091 (69) | 0 | 1090 | 1091 |
| VIP1 | CAGL0M09823g | 832/1113 (74) | 0 | 1147 | 1128 |
| PHO85 | CAGL0L12474g | 273/299 (91) | 3E-159 | 306 | 302 |
| PHO81 | CAGL0L06622g | 344/900 (38) | 5E-144 | 1179 | 1137 |
| ADK1 | CAGL0K11418g | 201/222 (90) | 6E-113 | 223 | 222 |
| PHO23 | CAGL0G06556g | 197/336 (58) | 5E-88 | 331 | 316 |
| PHO86 | CAGL0L05456g | 163/300 (54) | 5E-86 | 312 | 306 |
| PHO2 | CAGL0L07436g | 180/384 (46) | 8E-85 | 560 | 515 |
| PHO80 | CAGL0E02541g | 138/261 (52) | 7E-73 | 294 | 342 |
| PHO88 | CAGL0K12276g | 134/196 (68) | 8E-68 | 189 | 194 |
| PHO4 | CAGL0D05170g | 32/60 (53) | 4E-08 | 313 | 533 |
| SPL2 | No ortholog | ||||
| PHO89 | No ortholog | ||||
| NCP1 | CAGL0D04114g | 457/693 (65) | 0 | 692 | 687 |
| PHM1 | CAGL0F02145g | 499/831 (60) | 0 | 829 | 816 |
| PHM2 | CAGL0F02145g | 489/854 (57) | 0 | 836 | 816 |
| PHM3 | CAGL0G06952g | 582/719 (80) | 0 | 722 | 717 |
| PHO8 | CAGL0H07359g | 322/488 (65) | 4E-176 | 567 | 545 |
| AUT4 | CAGL0B00770g | 311/538 (57) | 1E-162 | 529 | 541 |
| PHO91 | CAGL0I05632g | 289/690 (41) | 8E-133 | 895 | 886 |
| SDT1 | CAGL0H09218g | 158/265 (59) | 6E-90 | 281 | 279 |
| IRC15 | CAGL0F01947g | 183/471 (38) | 5E-79 | 500 | 493 |
| GIT1 | CAGL0A01243g | 146/483 (30) | 3E-60 | 519 | 531 |
| PHM8 | CAGL0C02321g | 136/292 (46) | 2E-58 | 322 | 314 |
| PHM4 | CAGL0M12705g | 110/124 (88) | 1E-53 | 130 | 127 |
| CTF19 | CAGL0F02035g | 105/334 (31) | 6E-32 | 370 | 373 |
| PHM6 | No ortholog | ||||
| PHO5 | No ortholog | ||||
| PHO11 | No ortholog | ||||
| PHO12 | No ortholog | ||||
. | . | Sc to Cg . | Length of protein . | ||
|---|---|---|---|---|---|
| Gene . | Cg systematic name . | Identity (%) . | Expect . | Sc . | Cg . |
| MSN5 | CAGL0M01144g | 935/1219 (76) | 0 | 1225 | 1221 |
| PHO84 | CAGL0B02475g | 448/586 (76) | 0 | 588 | 580 |
| PHO87 | CAGL0F02387g | 649/966 (67) | 0 | 924 | 952 |
| PHO90 | CAGL0F02387g | 624/957 (65) | 0 | 882 | 952 |
| PSE1 | CAGL0M13871g | 754/1091 (69) | 0 | 1090 | 1091 |
| VIP1 | CAGL0M09823g | 832/1113 (74) | 0 | 1147 | 1128 |
| PHO85 | CAGL0L12474g | 273/299 (91) | 3E-159 | 306 | 302 |
| PHO81 | CAGL0L06622g | 344/900 (38) | 5E-144 | 1179 | 1137 |
| ADK1 | CAGL0K11418g | 201/222 (90) | 6E-113 | 223 | 222 |
| PHO23 | CAGL0G06556g | 197/336 (58) | 5E-88 | 331 | 316 |
| PHO86 | CAGL0L05456g | 163/300 (54) | 5E-86 | 312 | 306 |
| PHO2 | CAGL0L07436g | 180/384 (46) | 8E-85 | 560 | 515 |
| PHO80 | CAGL0E02541g | 138/261 (52) | 7E-73 | 294 | 342 |
| PHO88 | CAGL0K12276g | 134/196 (68) | 8E-68 | 189 | 194 |
| PHO4 | CAGL0D05170g | 32/60 (53) | 4E-08 | 313 | 533 |
| SPL2 | No ortholog | ||||
| PHO89 | No ortholog | ||||
| NCP1 | CAGL0D04114g | 457/693 (65) | 0 | 692 | 687 |
| PHM1 | CAGL0F02145g | 499/831 (60) | 0 | 829 | 816 |
| PHM2 | CAGL0F02145g | 489/854 (57) | 0 | 836 | 816 |
| PHM3 | CAGL0G06952g | 582/719 (80) | 0 | 722 | 717 |
| PHO8 | CAGL0H07359g | 322/488 (65) | 4E-176 | 567 | 545 |
| AUT4 | CAGL0B00770g | 311/538 (57) | 1E-162 | 529 | 541 |
| PHO91 | CAGL0I05632g | 289/690 (41) | 8E-133 | 895 | 886 |
| SDT1 | CAGL0H09218g | 158/265 (59) | 6E-90 | 281 | 279 |
| IRC15 | CAGL0F01947g | 183/471 (38) | 5E-79 | 500 | 493 |
| GIT1 | CAGL0A01243g | 146/483 (30) | 3E-60 | 519 | 531 |
| PHM8 | CAGL0C02321g | 136/292 (46) | 2E-58 | 322 | 314 |
| PHM4 | CAGL0M12705g | 110/124 (88) | 1E-53 | 130 | 127 |
| CTF19 | CAGL0F02035g | 105/334 (31) | 6E-32 | 370 | 373 |
| PHM6 | No ortholog | ||||
| PHO5 | No ortholog | ||||
| PHO11 | No ortholog | ||||
| PHO12 | No ortholog | ||||
Orthologs of the PHO pathway components, both regulators and transcriptional output, in S. cerevisiae (Sc) were determined for C. glabrata (Cg) on the basis of sequence similarity. Components are listed from lowest expect value to highest expect value and components with no ortholog are listed last. MSN5 through PHO89 are signaling components and NCP1 through PHO12 are genes upregulated in response to phosphate starvation.
Detection of phosphatase activity:
For a semiquantitative phosphatase assay, the agar plates with colonies were overlaid with α-naphthyl phosphate, Fast Blue Salt B stain, and 0.1 m sodium acetate (pH 4.2) (Wykoff et al. 2007). This assay causes a colony to turn red when phosphatase is secreted and remain white in the absence of phosphatase. For quantification of p-nitrophenyl phosphatase activity, strains were grown in high or no phosphate conditions in liquid media for 16 hr, with high phosphate samples diluted to stay in logarithmic phase. One mL of cells (OD600 ∼0.5) was pelleted and resuspended in sterile water. Measured units of phosphatase activity were expressed as OD400/OD600 (Huang and O'Shea 2005). Data were normalized to either wild type grown in phosphate starvation or pho4Δ + PHO4 such that induction for these strains was 100% (maximal induction expected).
Quantitative reverse-transcription PCR:
RNA was extracted by a standard phenol–chloroform protocol (Huang and O'Shea 2005). RNA was converted to cDNA with a reverse-transcription reaction (BIO-RAD iScript cDNA synthesis kit). Quantitative PCR using a 50-μl PCR reaction with Sybr Green I (Sigma-Aldrich, St. Louis) was performed. Primers were designed for ACT1, PHO84, and PHO5 for S. cerevisiae and ACT1, PHO84, and GIT1 for C. glabrata (see Table S1). Data were normalized to expression of ACT1 to control for loading differences, and we confirmed that ACT1 transcript abundance does not change dramatically during phosphate starvation (data not shown). These values were then normalized as described above to 100% induction.
Immunoblot analysis:
Abundance of S. cerevisiae and C. glabrata Pho4 protein was analyzed by immunoblot. The pho4Δ + PHO4 strains for both species were grown in high- and no-phosphate conditions as described. Protein was extracted and quantified and 30 μg of protein were subjected to SDS–PAGE and analyzed as described previously (Wykoff and O'Shea 2005).
RESULTS
Comparative genomic analysis of PHO pathway in S. cerevisiae and C. glabrata:
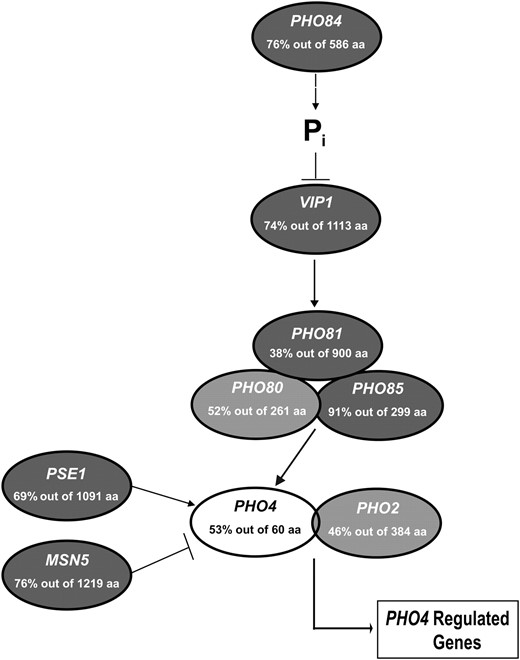
To determine PHO pathway conservation in C. glabrata, we identified orthologous sequences for each component of the pathway using BLASTP and the yeast genome order browser (Byrne and Wolfe 2005). Utilizing a predetermined cutoff value of E = 10−2, most of the components of the PHO pathway in S. cerevisiae have clear orthologs in C. glabrata (Figure 1 and Table 2), suggesting that a PHO pathway of similar architecture is present. However, C. glabrata does not contain orthologs of the acid phosphatases present in S. cerevisiae (PHO5, PHO11, and PHO12) or the Pho89 high-affinity phosphate transporter, nor is there obvious positive feedback through SPL2 (Persson et al. 2003; Wykoff et al. 2007). It is unclear what genes encode the inducible acid phosphatase activity because there are no homologs of acid phosphatases in the C. glabrata genome, but there is clear inducible acid phosphatase activity (Figure 2A). The transcription factor Pho4 is the least conserved component of the pathway and is identifiable by amino acid identity in the C-terminal DNA binding domain (Figure S1).
Genomic sequence predicts a C. glabrata PHO pathway similar to S. cerevisiae. The values indicate the percentage of amino acid identity and the number of amino acids over which the identity was measured by BLASTP alignment. Shading of each component differs on the basis of the expect (E) value for each C. glabrata ortholog. Components with an E value <10−100 are darkly shaded; E values >10−100 are lightly shaded; and E values >10−10 are open. Systematic C. glabrata names are given in Table 2.
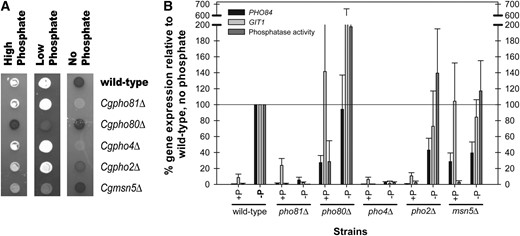
C. glabrata mutants defective in the PHO pathway components have similar phenotypes as S. cerevisiae mutants, except for Cgpho2Δ. (A) Acid phosphatase activity of C. glabrata mutants defective in major PHO pathway components. Strains were grown in high (SD), low (YEPD), and phosphate starvation (SD no phosphate) conditions and solid media was overlaid with substrate to detect acid phosphatase activity. Dark shaded colonies have acid phosphatase activity and white colonies have reduced phosphatase activity. (B) Induction of PHO84 and GIT1 and phosphatase activity of C. glabrata mutants. Transcription of PHO84 and GIT1 was measured with reverse-transcription quantitative PCR. Data were normalized to wild-type cells grown in phosphate starvation such that expression for this strain was 100% (maximal induction expected during starvation). p-nitrophenyl phosphatase activity was quantified and normalized to wild type. Data for PHO84 and GIT1 are representative of four independent experiments and data for phosphatase activity are representative of three independent experiments. Standard error was calculated for PHO84, GIT1, and phosphatase activity. The value for GIT1 in pho80Δ without phosphate is 380 ± 279%. The errors not shown are 63% for GIT1 in pho80Δ with phosphate, and 135% for phosphatase activity in pho80Δ without phosphate.
Confirmation of PHO pathway functions in C. glabrata:
To confirm that the identified orthologs regulate a phosphate-starvation-inducible acid phosphatase, we inactivated candidate genes with antibiotic resistance genes KANMX6 or NATMX6 (Longtine et al. 1998). Assaying these mutants by a semiquantitative assay for acid phosphatase activity, we confirmed that CgPho4 is required for induction of phosphatase activity during phosphate starvation (Figure 2A). We further determined that CgPho4 is regulated by the orthologous cyclin/CDK/CDK inhibitor complex and that CgPho4 localization is likely one mode of regulation as deletion of CgMSN5, a putative exporter of CgPho4, results in partial activation of the pathway. Notably, deletion of CgPHO2 does not abolish phosphatase induction, as it does in S. cerevisiae (Yoshida et al. 1989a; Springer et al. 2003).
Using quantitative PCR, we quantified the amount of transcript of two genes we hypothesized were regulated by extracellular phosphate status, CgPHO84 and CgGIT1, and confirmed that they are transcriptionally induced during phosphate starvation (Figure 2B). Quantitative analysis of the mutant strains supported the semiquantitative conclusions, although the two promoters behaved slightly differently from one another. Because the acid phosphatase gene has not been identified, we cannot directly compare transcriptional induction of these two genes with induction of phosphatase activity. Deletion of PHO4 in C. glabrata results in no increase in gene expression during phosphate starvation, which is true in S. cerevisiae. Unlike S. cerevisiae, C. glabrata exhibits a reduced dependence on the Pho2 transcription factor. Although not at wild-type levels, gene expression of the starvation-regulated promoters still occurs when PHO2 is deleted in C. glabrata.
C. glabrata Pho4 is sufficient for Pho2 independence:
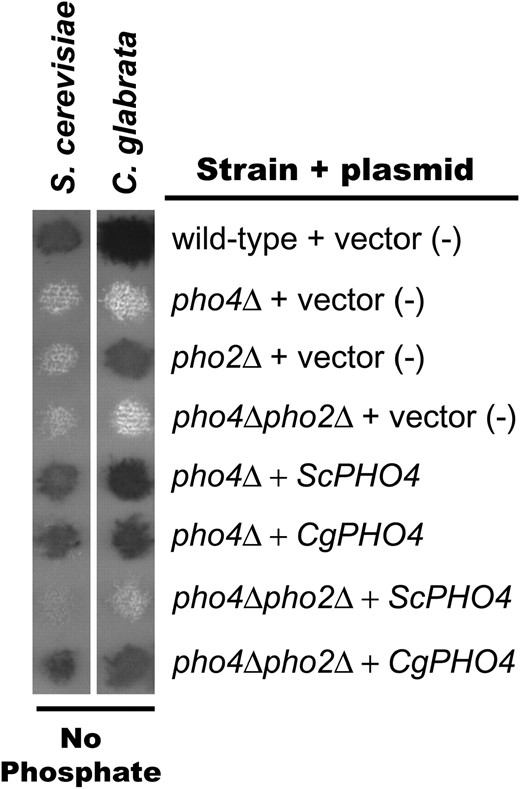
To determine whether the Pho2 independence in C. glabrata is due to alterations in the C. glabrata Pho4 transcription factor (in trans) or alterations to C. glabrata promoters (in cis), we characterized the ability of Pho4, from both S. cerevisiae and C. glabrata, to induce gene expression independent of Pho2 in either species. Plasmids containing PHO4 or PHO2 genes from either S. cerevisiae or C. glabrata were generated and transformed into S. cerevisiae and C. glabrata mutants defective in one or both transcription factors (pho4Δ, pho2Δ, or pho4Δpho2Δ). PHO4 from either species complemented or cross-complemented pho4Δ strains when subjected to the semiquantitative phosphatase assay, suggesting the plasmids contain functional genes (Figure 3). We also confirmed cross-complementation with PHO2, although CgPHO2 complemented S. cerevisiae to a lower extent than the converse condition (Figure 4, A and B and Figure S2). We confirmed our semiquantitative data by quantifying the amount of ScPHO84 and ScPHO5 transcript in S. cerevisiae strains (Figure 4A) and the amount of CgPHO84 and CgGIT1 transcript in C. glabrata strains (Figure 4B).
Semiquantitative phosphatase assay demonstrating that PHO4 plasmids are functional and CgPHO4 is sufficient for Pho2 independence. S. cerevisiae and C. glabrata mutants lacking one or both transcription factors (pho4Δ, pho2Δ, or pho4Δpho2Δ) contain either empty vector (pRS313), ScPHO4, or CgPHO4 plasmids. These strains were grown on solid media lacking phosphate and overlaid with phosphatase substrate.
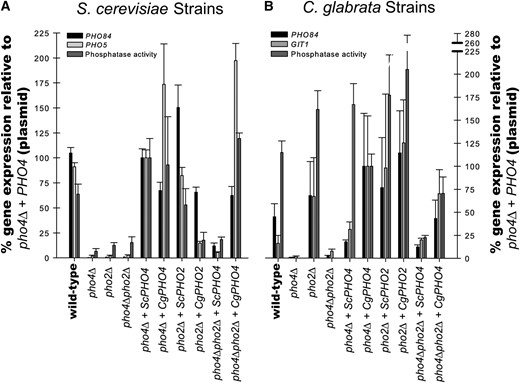
Quantification of phosphate responsive genes in S. cerevisiae strains and C. glabrata strains. (A) S. cerevisiae mutants lacking one or both transcription factors (pho4Δ, pho2Δ, or pho4Δpho2Δ) were generated to contain either empty vector (pRS313), ScPHO4, CgPHO4, ScPHO2, or CgPHO2 plasmids. These strains were grown in media lacking phosphate and quantitative reverse-transcription PCR was used to measure amount of PHO84 and PHO5 transcript. Data were normalized to Scpho4Δ + ScPHO4 (such that expression for this strain was 100%) rather than to wild type because the plasmid alters the copy number of the PHO4. p-nitrophenyl phosphatase activity was also normalized to Scpho4Δ + ScPHO4. (B) C. glabrata mutants lacking one or both transcription factors with the same plasmids as in A. These strains were treated as described for S. cerevisiae strains except that induction of GIT1 was measured for C. glabrata rather than PHO5. Data were normalized to Cgpho4Δ + CgPHO4 for both the quantitative reverse-transcription PCR and the phosphatase assay. The standard error is of at least three independent replicates.
We hypothesized that if Pho2 dependence is a consequence of alterations in the Pho4 protein, then only CgPho4 should induce phosphate starvation genes in the pho4Δpho2Δ mutant in both S. cerevisiae and C. glabrata. However, if the Pho2 dependence is due to changes in the promoter regions (cis-regulatory regions), then both ScPho4 and CgPho4 should induce starvation genes in the C. glabrata pho4Δpho2Δ, but neither ScPho4 nor CgPho4 would induce transcription in the S. cerevisiae pho4Δpho2Δ. Finally, if the Pho2 dependence is due to changes in both the Pho4 protein and the promoter regions, then both ScPho4 and CgPho4 should induce starvation genes in C. glabrata pho4Δpho2Δ and only CgPho4 should induce starvation genes in S. cerevisiae pho4Δpho2Δ.
Analysis of gene expression during phosphate starvation demonstrates that only CgPho4, and not ScPho4, can dramatically induce starvation genes in the absence of the Pho2 in either species (Figure 4, A and B). Specifically, CgPho4 suppresses the Pho2 dependence in S. cerevisiae and ScPho4 is not sufficient in Cgpho4Δpho2Δ. These results suggest that the Pho2 dependence is a consequence of alterations in the Pho4 protein and that CgPho4 obviates the need for Pho2. The CgPho4 generation of Pho2 independence is most clear in S. cerevisiae strains; with CgPho4, the S. cerevisiae promoters do not require Pho2. While not as dramatic, C. glabrata promoters exhibit similar behavior with ScPho4 being dependent on Pho2 and CgPho4 not requiring Pho2, but we cannot eliminate the possibility that there are subtle cis changes to phosphate-regulated promoters.
While Pho2 may serve an ancillary role in C. glabrata and the promoters in C. glabrata have a low affinity for CgPho2, ScPho4 must recruit CgPho2 to some C. glabrata promoters because ScPho4 is functional in a Cgpho4Δ background. Because the consensus binding site for Pho2 is an A/T-rich region (Barbaric et al. 1996), we are not able to determine whether there are Pho2 binding sites within the phosphate-regulated promoters.
C. glabrata likely confers Pho2 independence as a consequence of its increased size and not because of overexpression:
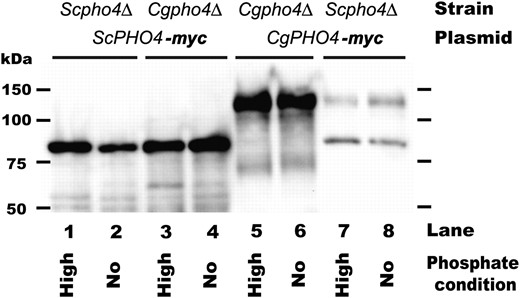
Previous studies have demonstrated that when Pho4 is overexpressed in S. cerevisiae (∼20-fold higher than wild-type expression), transcription of PHO5 can be induced in the absence of Pho2, although expression is only ∼25% of wild-type levels (Yoshida et al. 1989a,b; Barbaric et al. 1996). This situation appears to resemble that of a Cgpho2Δ strain, where phosphate-regulated promoters are induced, but less than wild-type phosphate starvation levels. To determine whether the Pho2 independence in C. glabrata is a consequence of overexpression of the PHO4 during phosphate starvation conditions, we examined the abundance of the S. cerevisiae and C. glabrata Pho4 proteins in high- and no-phosphate conditions by immunoblot analysis (Figure 5).
Immunoblot analysis demonstrating little change in Pho4 abundance between high- and no-phosphate conditions. S. cerevisiae and C. glabrata pho4Δ strains with a PHO4 plasmid with 13-myc epitope were grown to log phase, washed, and then transferred to high- and no-phosphate conditions for 3 hr. Pho4 was detected with 9E10 as described previously (Wykoff and O'Shea 2005). The expected sizes of Pho4 are 54 kDa in S. cerevisiae and 79 kDa in C. glabrata (size of Pho4 + 13-myc tag); however, previous studies demonstrate ScPho4 migrates ∼25 kDa heavier than expected and has a degradation product (Kaffman et al. 1998; O'Neill et al. 1996), and CgPHO4 also appears to migrate aberrantly. This immunoblot is representative of three independent experiments.
We demonstrate that neither ScPho4 nor CgPho4 change abundance dramatically in high- and no-phosphate conditions. Although CgPho4 is much fainter for Scpho4Δ + CgPHO4, the amount of protein is still the same for both phosphate conditions. The lack of abundance changes is unlikely to be an artifact of C-terminally tagging the proteins with a myc epitope because the myc-tagged genes complement many deletion strains (Figures 3 and 4). These results suggest that the Pho2 independence is not a consequence of overexpression of CgPho4.
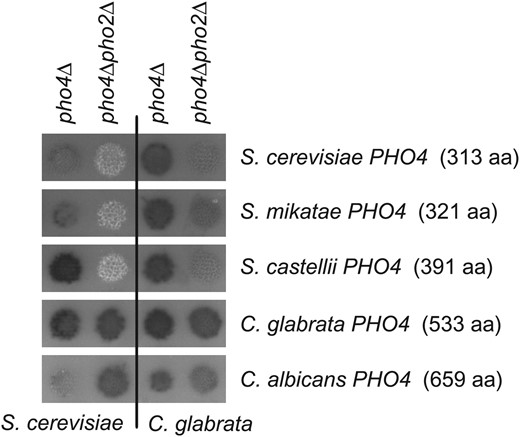
Our results demonstrate that only CgPho4, and not ScPho4, can suppress the Pho2 dependence in either species. This difference prompted us to look at the difference between the Pho4 transcription factors from the two species. One major difference is the size of the Pho4 proteins: C. glabrata Pho4, which is 533 amino acids, is much larger than S. cerevisiae Pho4, which is only 313 amino acids. Of all of the orthologs that we identified between the two species, this is the greatest difference in size (Table 2). An examination of the ascomycete lineage reveals that the Pho4 protein in the Saccharomyces genus is small relative to all of the other sequenced ascomycetes. Furthermore, an extensive analysis of Neurospora crassa mutants identified many shared components of the PHO pathway, except that NcPHO2 was never demonstrated to be important for phosphatase induction during phosphate starvation (Peleg et al. 1996a,b; Yang et al. 1991). We hypothesized that the Pho2 dependence is a trait that has only appeared in species closely related to S. cerevisiae with relatively small Pho4 proteins.
To begin testing this hypothesis, S. cerevisiae and C. glabrata mutants defective in one or both transcription factors (pho4Δ and pho4Δpho2Δ) were transformed with plasmids containing PHO4 from various ascomycetes (Figure 6). These strains were tested for phosphatase activity. The pho4Δ mutants into which PHO4 plasmids were transformed demonstrate that the plasmids contain functional genes; the defect caused by the deletion of ScPHO4 or CgPHO4 is removed and phosphatase activity is detected. The results for the pho4Δ pho2Δ mutants with PHO4 plasmids resemble the data obtained for ScPho4 and CgPho4 in the pho4Δ pho2Δ mutants. Only the larger Pho4 transcription factors [i.e., C. glabrata (533 aa) and C. albicans (659 aa)] induce phosphatase activity independent of Pho2 in both species. The smaller Pho4 transcription factors [i.e., S. cerevisiae (313 aa), S. mikatae (321 aa), S. castellii (391 aa)], however, do not induce the phosphatase in the absence of Pho2. These results suggest that Pho2 independence is likely a consequence of the increased size of the Pho4 transcription factor.
Phosphatase activity in S. cerevisiae and C. glabrata mutants containing the Pho4 transcription factor from various hemiascomycetes. Plasmids containing the PHO4 gene from various hemiascomycetes (labeled on the right) were transformed into S. cerevisiae (left two columns) and C. glabrata mutants (right two columns) defective in one or both transcription factors (pho4Δ and pho4Δpho2Δ). Phosphatase activity was determined as described in Figure 2 and is representative of multiple transformants.
DISCUSSION
We demonstrate that genomic sequence is highly predictive of the PHO signal transduction pathway in C. glabrata. Through mutant analysis, CgPho4 is likely regulated by the cyclin/CDK/CDK inhibitor complex Pho80/Pho85/Pho81 and likely regulated by localization in the same fashion as S. cerevisiae. This is supported by deletion of PHO81, which is unable to induce phosphatase expression during phosphate starvation in both S. cerevisiae and C. glabrata, and deletion of PHO80 resulting in a constitutive phenotype. It is unlikely that these phenotypes are independent of the kinase activity of this complex. Furthermore, the known phosphorylation sites of ScPho4 are moderately conserved in CgPho4 also suggesting that these sites are used in C. glabrata (Figure S1). Finally, the expression of the phosphatase is increased in high phosphate conditions in a Cgmsn5Δ strain, identical to Scmsn5Δ strains. Whereas we have not directly demonstrated that CgPho4 is a substrate for the kinase complex, the simplest explanation is that CgPho4 is regulated in a similar fashion to S. cerevisiae.
Switching Pho4 between species demonstrates that CgPho4 is necessary and sufficient for Pho2-independent induction of phosphate starvation-regulated promoters in both species. The Pho2 independence of PHO promoters in C. glabrata was an unexpected result and highlights the need for studies in nonmodel species. Interestingly, there are additional differences between the PHO pathway of S. cerevisiae and C. glabrata, including the lack of a known acid phosphatase gene and the relative dearth of canonical Pho4 binding sites (CACGTG) in many putative phosphate-regulated genes in C. glabrata (data not shown), suggesting that there are many subtle differences in how the two organisms regulate their phosphate starvation response.
The view of speciation being influenced by subtle changes to promoters has been disputed by many recent studies and our study supports the idea that species have exploited changes in trans in the PHO pathway of ascomycetes (King and Wilson 1975; Piano et al. 1999; Wittkopp et al. 2004, 2008; Hoekstra et al. 2006; Chang et al. 2008). On the basis of size and conservation of Pho4 in ascomycetes that did not experience a whole genome duplication event, we hypothesize that Pho2 independence is an ancestral trait, which is supported by small Pho4 proteins appearing to be Pho2 dependent. Because of the relative promiscuousness of Pho2 binding (it binds to A/T-rich sequences of DNA) (Barbaric et al. 1996; Zhang et al. 1997), we cannot exclude the possibility that promoter changes have occurred, but clearly the Pho2 independence depends on domains within CgPho4. We expect that the Saccharomyces species that appear Pho2 dependent likely experienced a selective advantage because of this increased pathway complexity.
C. glabrata, as a commensal pathogen of mammals (Redondo-Lopez et al. 1990; Cormack and Falkow 1999), likely experiences two inorganic phosphate conditions: high phosphate conditions during growth in mammalian tissues and extremely low phosphate conditions on external epithelium. S. cerevisiae, however, likely experiences a different condition: growth on decaying organic matter that might provide organic phosphates and moderate levels of inorganic phosphate. Contrasting these niches provides for a possible selective advantage for Saccharomyces species. During conditions of low inorganic phosphate, S. cerevisiae through Pho4 is able to induce a subset of phosphate starvation responsive genes, including the high-affinity phosphate transporter PHO84, but maintain relative repression of the acid phosphatase gene PHO5 (Springer et al. 2003). This alternate gene regulation program results from an isoform of Pho4 that is unphosphorylated except on one site (Springer et al. 2003). Phosphorylation of serine 223 decreases the interaction between Pho4 and Pho2 (O'Neill et al. 1996; Komeili and O'Shea 1999). Because the acid phosphatase gene in C. glabrata is unidentified, we are unable to determine whether this same intermediate phosphate starvation response occurs in C. glabrata, but future studies will explore this question.
We observe a different signaling architecture in the PHO pathway of C. glabrata relative to budding yeast and others have observed differences between pathways such as galactose metabolism and mating pathways between the two species (Tsong et al. 2003; Butler et al. 2004; Hittinger et al. 2004). The two species inhabit different environments and require divergent responses for optimal growth. It is appealing to hypothesize that trans-regulatory changes to signaling pathways have allowed the two different species to exploit their environmental niches through changes in the transcription factors that regulate entire gene expression programs.
Footnotes
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.101063/DC1.
Footnotes
Communicating editor: A. P. Mitchell
Acknowledgements
We thank Rochelle Argentieri for optimization of the C. glabrata qPCR primers, Heather Eberhart for collection of mutant qPCR data, and Brendan Cormack for yeast strains. We thank Erin K. O'Shea, who generously allowed for preliminary experiments to be performed in her laboratory. We thank Jonathan Raser, Narendra Maheshri, and Erin K. O'Shea for insightful comments and suggestions on the manuscript. This work was funded by the Department of Biology at Villanova University and by a grant from the National Science Foundation (RUI-MCB-0747799).
References
Alon, U., M. G. Surette, N. Barkai and S. Leibler,
Barbaric, S., M. Munsterkotter, J. Svaren and W. Horz,
Butler, G., C. Kenny, A. Fagan, C. Kurischko, C. Gaillardin et al.,
Byrne, K. P., and K. H. Wolfe,
Carroll, A. S., A. C. Bishop, J. L. DeRisi, K. M. Shokat and E. K. O'Shea,
Cecilia Garofalo, E. S.,
Chang, Y. W., F. G. Robert Liu, N. Yu, H. M. Sung, P. Yang et al.,
Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton et al.,
Cormack, B. P., and S. Falkow,
Cormack, B. P., N. Ghori and S. Falkow,
Domergue, R., I. Castano, A. De Las Penas, M. Zupancic, V. Lockatell et al.,
Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola et al.,
Fry, R. C., M. S. DeMott, J. P. Cosgrove, T. J. Begley, L. D. Samson et al.,
Gasch, A. P.,
Guthrie, C., and G. R. Fink,
Hentges, P., B. Van Driessche, L. Tafforeau, J. Vandenhaute and A. M. Carr,
Hittinger, C. T., A. Rokas and S. B. Carroll,
Hoekstra, H. E., R. J. Hirschmann, R. A. Bundey, P. A. Insel and J. P. Crossland,
Huang, S., and E. K. O'Shea,
Ihmels, J., S. Bergmann, M. Gerami-Nejad, I. Yanai, M. McClellan et al.,
Kaffman, A., N. M. Rank, E. M. O'Neill, L. S. Huang and E. K. O'Shea,
King, M. C., and A. C. Wilson,
Komeili, A., and E. K. O'Shea,
Lenburg, M. E., and E. K. O'Shea,
Longtine, M. S., A. McKenzie, 3rd, D. J. Demarini, N. G. Shah, A. Wach et al.,
Migeon, B. R., J. Axelman and P. Jeppesen,
Moses, A. M., D. A. Pollard, D. A. Nix, V. N. Iyer, X. Y. Li et al.,
O'Neill, E. M., A. Kaffman, E. R. Jolly and E. K. O'Shea,
Oshima, Y.,
Peleg, Y., R. Addison, R. Aramayo and R. L. Metzenberg,
Peleg, Y., R. Aramayo, S. Kang, J. G. Hall and R. L. Metzenberg,
Persson, B. L., J. O. Lagerstedt, J. R. Pratt, J. Pattison-Granberg, K. Lundh et al.,
Piano, F., M. J. Parisi, R. Karess and M. P. Kambysellis,
Pinter, R. Y., O. Rokhlenko, E. Yeger-Lotem and M. Ziv-Ukelson,
Rao, C. V., J. R. Kirby and A. P. Arkin,
Redondo-Lopez, V., M. Lynch, C. Schmitt, R. Cook and J. D. Sobel,
Schneider, K. R., R. L. Smith and E. K. O'Shea,
Sikorski, R. S., and P. Hieter,
Simon, J. A.,
Springer, M., D. D. Wykoff, N. Miller and E. K. O'Shea,
Tsong, A. E., M. G. Miller, R. M. Raisner and A. D. Johnson,
Vogel, K., W. Horz and A. Hinnen,
Wittkopp, P. J., B. K. Haerum and A. G. Clark,
Wittkopp, P. J., B. K. Haerum and A. G. Clark,
Wolfe, K. H.,
Wykoff, D. D., and E. K. O'Shea,
Wykoff, D. D., and E. K. O'Shea,
Wykoff, D. D., A. H. Rizvi, J. M. Raser, B. Margolin and E. K. O'Shea,
Yang, J. W., S. S. Dhamija and M. E. Schweingruber,
Yeger-Lotem, E., S. Sattath, N. Kashtan, S. Itzkovitz, R. Milo et al.,
Yoshida, K., Z. Kuromitsu, N. Ogawa and Y. Oshima,
Yoshida, K., N. Ogawa and Y. Oshima,
Zhang, F., M. Kirouac, N. Zhu, A. G. Hinnebusch and R. J. Rolfes,