-
PDF
- Split View
-
Views
-
Cite
Cite
Yubin Li, Hugo K Dooner, Excision of Helitron Transposons in Maize, Genetics, Volume 182, Issue 1, 1 May 2009, Pages 399–402, https://doi.org/10.1534/genetics.109.101527
Close - Share Icon Share
Abstract
Helitrons are novel transposons discovered by bioinformatic analysis of eukaryotic genome sequences. They are believed to move by rolling circle (RC) replication because their predicted transposases are homologous to those of bacterial RC transposons. We report here evidence of somatic Helitron excision in maize, an unexpected finding suggesting that Helitrons can exhibit an excisive mode of transposition.
HELITRONS constitute a major superfamily of transposons discovered recently by a computational analysis of genomic sequences from Arabidopsis, rice, and Caenorhabditis elegans (Kapitonov and Jurka 2001). These presumed class II elements, unlike most other elements in that class, are postulated to transpose by a copy-and-paste mechanism. The absence of target-site duplications and the homology of the putative transposase encoded by the “consensus” autonomous element to the transposase of bacterial rolling circle (RC) transposons (Mendiola and de la Cruz 1992) led Kapitonov and Jurka (2001) to postulate that Helitrons transposed by an RC replication, rather than by an excision-repair, mechanism. Helitrons appear to be ubiquitous components of eukaryotic genomes, as they have been found in organisms ranging from fungi to vertebrates, including mammals (Poulter et al. 2003; Pritham and Feschotte 2007). However, there is at present no evidence for an autonomous Helitron element or a transposition mechanism in any organism.
Helitrons have few constant structural features. They have conserved 5′-TC and CTRR-3′ termini, carry a 16- to 20-bp palindrome of variable sequence ∼10–12 bp upstream of the 3′ terminus, and insert invariably between host nucleotides A and T. The reconstructed putative autonomous elements in Arabidopsis and rice are large (5.5–15 kb) and encode proteins with homology to a DNA helicase and an RPA-like, single-stranded DNA-binding protein (Kapitonov and Jurka 2001). A recently described 11.5-kb Helitron in Ipomoea carries both of these genes, but with prematurely terminating mutations in each (Choi et al. 2007). The vast majority of Helitrons in Arabidopsis and C. elegans are shorter nonautonomous elements that do not encode proteins. Like other transposons, Helitrons can be mutagenic and have been reported in spontaneous mutations in maize (Lal et al. 2003; Gallavotti et al. 2004; Chuck et al. 2007) and in morning glory (Choi et al. 2007). The mutagenic maize Helitrons are large and have captured fragments of several unrelated genes. The Helitrons that contribute to the apparent breakdown in genetic colinearity among bz haplotypes (Fu and Dooner 2002) are also large and carry sequences from yet other genes (Lai et al. 2005; Morgante et al. 2005). In fact, the diversity of element complement is greater in maize than in other plant species (Hollister and Gaut 2007; Sweredoski et al. 2008). Smaller Helitrons lacking gene fragments and resembling the agenic Helitrons of Arabidopsis and C. elegans were recently identified in maize from a vertical comparison of 8 bz haplotypes (Wang and Dooner 2006). In spite of the extensive Helitron insertion polymorphisms found in maize and other species, an actual Helitron transposition event has yet to be reported.
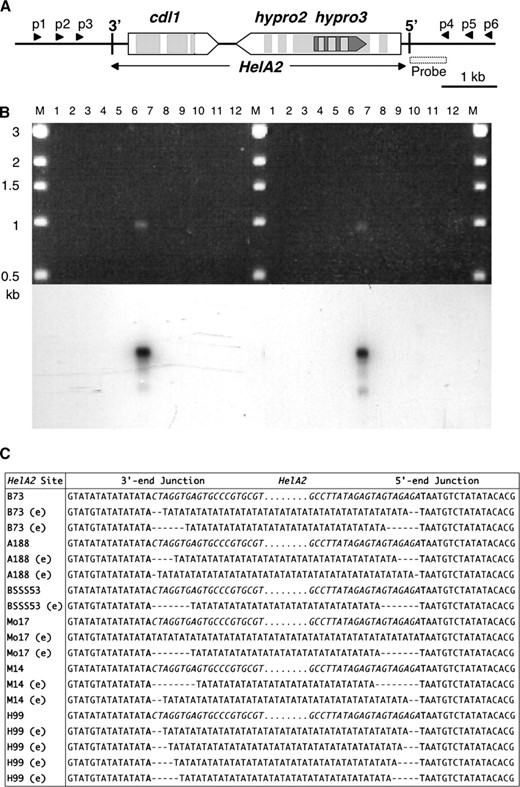
Somatic excision of the 6.0-kb HelA2 Helitron in 5S:
Several Helitrons are polymorphic among 9S bz haplotypes (Wang and Dooner 2006). The simplest explanation for the +/− polymorphism is that the unoccupied site was never visited. Nevertheless, because the actual mechanism of transposition of Helitrons is not known, the site lacking a Helitron has been referred to as “vacant” to distinguish it from the footprint-bearing “empty sites” produced by the excision of most class II DNA transposons (Lai et al. 2005). Helitron HelA1 is 5.8 kb long, contains sequences for three genes, and is found only in the McC and W22 bz haplotypes (Dooner and He 2008). A nearly identical copy of HelA1, designated HelA2, is found in 5S in the inbred lines B73, McC, W22, A188, A636, B73, H99, M14, Mo17, and BSSS53, but not in W23. Thus, most of the Corn Belt lines examined lack the HelA copy in 9S and have only one in 5S (Lai et al. 2005; Wang and Dooner 2006).
RC transposons are not supposed to excise (del Pilar Garcillan-Barcia et al. 2002), so lines carrying HelA2 in 5S are not expected to produce PCR products equivalent to transposon “empty” sites. We tested this prediction in DNA from different inbreds by performing nested PCR with various combinations of primers flanking HelA2 (Figure 1A). Unexpectedly, we obtained PCR products of a size similar to excision empty sites in some of the inbreds that carry HelA2 in 5S. One set of results is shown in Figure 1B, where M14 displays an ∼1-kb empty-site-sized PCR band that also hybridizes to a probe from a HelA2-adjacent sequence present in single copy in the maize genome (data not shown). Other inbreds, e.g., A188 and B73, gave empty-site-sized bands with other primer combinations. This difference among inbreds could reflect variability in HelA2 excision activity and/or polymorphisms for the primer sites, which were based on the sequence of B73 (Lai et al. 2005).
Evidence of HelA2 excision in 5S. (A) HelA2 structure in B73, showing the location of primers p1–p6 used for PCR. The exons and introns of the cdl, hypro2, and hypro3 gene fragments are open and shaded, respectively, pointing in their direction of transcription. (B, top) Agarose gel stained with ethidium bromide. HelA2 excision was tested by nested PCR on leaf DNA with the primer combinations p2/p6 and p3/p4 (left) or p1/p5 and p3/p4 (right). Lane 1, A188; lane 2, A636; lane 3, B73; lane 4, BSSS53; lane 5, H99; lane 6, M14; lane 7, Mo17; lane 8, 4Co63; lane 9, W23; lane 10, W22; lane 11, McC; lane 12, BAC b0511l12; lane M, DNA markers. The band in inbred M14 has the expected size of a Helitron excision product. (B, bottom) Hybridization of gel to a probe adjacent to HelA2. (C) Sequence of HelA2 full and empty (e) sites. Only part of the HelA2 3′ and 5′ junctions is shown, with the body of HelA2 indicated by dots. A sample of excision footprints from different inbreds is shown beneath the respective full sites. All excision footprints consist of a variable number of TA repeats (16–24). The AT host dinucleotide at the HelA2 insertion site is in boldface type and the HelA2 termini are in italic type (dashes introduced for alignment). No variability in TA repeat number was seen when multiple clones of a wild-type vacant site were sequenced (supporting information, Figure S1), ruling out in vitro DNA replication artifacts.
To investigate the nature of these putative excision products, the PCR bands from various inbreds (B73, A188, BSSS53, Mo17, M14, and H99) were cloned and sequenced (Figure 1C). Interestingly, all HelA2 excision site footprints consisted of a variable number of TA repeats (16–24) at the previous site of HelA2 insertion. The sequence adjacent to HelA2 is TA-rich and consists of a string of (TA/G) repeats at the 3′-end. The sequences in Figure 1C are clearly empty sites derived from previous sites of HelA2 insertion because they differ from each other by the same SNPs that distinguish the parental HelA2 insertion sites (located beyond the sequences shown in Figure 1C). Oligo-TA stretches were also found recently adjacent to TAFT elements, novel transposons that do not appear to be related to Helitrons, but rather to elements of the Mutator superfamily (Wang and Dooner 2006). The meaning of this coincidence is unclear, but may have more to do with a general mechanism of double-strand break repair by the host than with the specifics of transposition of the elements because excision of Helitrons from insertion sites lacking flanking TAs did not generate TA repeats at the excision sites (see below).
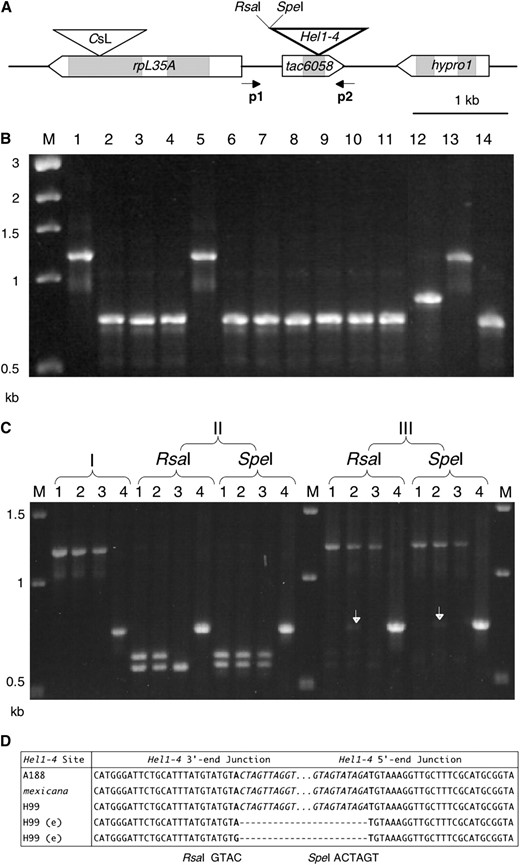
Somatic excision of the 0.4-kb Hel1-4 Helitron in 9S:
The short, 0.4-kb Hel1-4 element found in an intron of the single-copy tac6058 gene in the A188 bz haplotype (Wang and Dooner 2006) is present at the same location in inbred H99 and an accession of Chalco teosinte. Figure 2 shows PCR and sequence data documenting that Hel1-4 can excise from tac6058 in H99. Figure 2D presents the sequence of two empty sites, one without an excision footprint and one with a simple A-to-G transition at the “universal” AT dinucleotide insertion site. Polymorphisms beyond the tac6058 empty sites shown in Figure 2D were identical with those of H99. Thus, inbred H99 has the ability to excise both large and small Helitrons of the Hel1 family (Dooner et al. 2007).
Evidence of Hel1-4 excision in 9S. (A) Hel1-4 structure in the A188 bz haplotype, showing the location of primers p1 and p2 used for PCR. (B) Agarose gel stained with ethidium bromide. The presence or absence of Hel1-4 in a series of inbreds was tested on leaf DNA by PCR with primers 1 and 2. Lane 1, A188; lane 2, A636; lane 3, B73; lane 4, BSSS53; lane 5, H99; lane 6, M14; lane 7, Mo17; lane 8, 4Co63; lane 9, W23; lane 10, W22; lane 11, McC; lane 12, I137TN, lane 13, Zea mays ssp. mexicana; lane 14, Z. mays ssp. parviglumis. (C) Agarose gel stained with ethidium bromide. Lane 1, A188; 2, H99; 3, Zea mays ssp. mexicana; lane 4, McC (negative control). I, PCR amplification with p1 and p2 primers. II, PCR products from I were digested with RsaI and SpeI, which cut at the Hel1-4 3′ junction (A and D). RsaI also cuts within the mexicana Hel1-4 element, producing the polymorphic RsaI banding pattern in lane 3. No Hel1-4 excision products are seen if the genomic DNA is cut prior to PCR (data not shown). III, Detection of Hel1-4 excision by loss of restriction sites. The RsaI and SpeI digests from II were reamplified with primers p1 and p2. The bands indicated by the arrows in H99 represent Hel1-4 excision products. (D) Sequence of Hel1-4 full and empty (e) sites. Only part of the Hel1-4 3′ and 5′ junctions is shown, with the body of Hel1-4 indicated by dots. A sample of sequenced excision footprints from H99 is shown beneath the H99 full site. The AT host dinucleotide at the Hel1-4 insertion site is in boldface type and the Hel1-4 termini are in italic type (dashes introduced for alignment).
Somatic excision of a 2.8-kb Helitron from 10L:
Several putative maize Helitrons were identified recently in the GenBank nr database (Du et al. 2008). Primers flanking the putative Helitrons were used to analyze a panel of maize inbreds, and the PCR products were sequenced to identify +/− polymorphisms. One such polymorphism was identified for a 2.8-kb putative Helitron in 10L, with inbreds W22 and W23 lacking the insertion. In addition to the occupied site, B73 gave a faint empty-site-sized PCR product that lacked a footprint and was otherwise identical to the sequence flanking the insertion in the bacterial artificial chromosome (BAC) (Du et al. 2008). Lack of sequence polymorphisms in the adjacent sequences prevented the authors from excluding the possibility that this PCR product arose from DNA contamination in the B73 DNA preparation. However, considered against the data presented here, it is very likely that this band represents a Hel somatic excision product. Whether empty Helitron sites have or lack footprints may depend on the nature of the adjacent sequence.
Conclusions:
The data presented here clearly establish that Helitrons can excise somatically from their chromosomal sites, suggesting that, like Tn7 (Craig 2002) and Mutator (Walbot and Rudenko 2002), they may exhibit both replicative and excisive modes of transposition. Helitron excision activity was detected in some lines, but not in others, so it may be a polymorphic trait in maize. When present, the excision activity (excision band intensity) varies among lines. A screen for Helitron excision from an aleurone-pigmenting reporter construct would be valuable in tracking this activity. Two lines carrying Helitrons at more than one site, B73 and H99, produced empty sites for both, supporting the notion that the Helitron empty sites represent a general Helitron excision mechanism, rather than an unusual property of the adjacent sequences.
Footnotes
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.101527/DC1.
Footnotes
Communicating editor: J. A. Birchler
Acknowledgements
We are grateful to Jun Huang and Qinghua Wang for helpful comments on the manuscript. The work was supported by National Science Foundation grant DBI-03-20683.
References
Choi, J. D., A. Hoshino, K. I. Park, I. S. Park and S. Iida,
Chuck, G., R. Meeley, E. Irish, H. Sakai and S. Hake,
Craig, N. L.,
del Pilar Garcillan-Barcia, M., I. Bernales, M. V. Mendiola and F. de la Cruz,
Dooner, H. K., and L. He,
Dooner, H. K., S. K. Lal and L. C. Hannah,
Du, C., J. Caronna, L. He and H. K. Dooner,
Fu, H., and H. K. Dooner,
Gallavotti, A., Q. Zhao, J. Kyozuka, R. B. Meeley, M. K. Ritter et al.,
Hollister, J. D., and B. S. Gaut,
Kapitonov, V. V., and J. Jurka,
Lai, J., Y. Li, J. Messing and H. K. Dooner,
Lal, S. K., M. J. Giroux, V. Brendel, C. E. Vallejos and L. C. Hannah,
Mendiola, M. V., and F. de la Cruz,
Morgante, M., S. Brunner, G. Pea, K. Fengler, A. Zuccolo et al.,
Poulter, R. T., T. J. Goodwin and M. I. Butler,
Pritham, E. J., and C. Feschotte,
Sweredoski, M., L. DeRose-Wilson and B. S. Gaut,
Walbot, V., and G. N. Rudenko,
Wang, Q., and H. K. Dooner,