-
PDF
- Split View
-
Views
-
Cite
Cite
Emily J Kuhn-Parnell, Cecilia Helou, David J Marion, Brian L Gilmore, Timothy J Parnell, Marc S Wold, Pamela K Geyer, Investigation of the Properties of Non-gypsy Suppressor of Hairy-wing-Binding Sites, Genetics, Volume 179, Issue 3, 1 July 2008, Pages 1263–1273, https://doi.org/10.1534/genetics.108.087254
Close - Share Icon Share
Abstract
Insulators define interactions between transcriptional control elements in eukaryotic genomes. The gypsy insulator found in the gypsy retrovirus binds the zinc-finger Suppressor of Hairy-wing [Su(Hw)] protein that associates with hundreds of non-gypsy regions throughout the Drosophila genome. Models of insulator function predict that the gypsy insulator forms chromatin loop domains through interactions with endogenous Su(Hw) insulators (SIs) to limit the action of transcriptional control elements. Here we study SI 62D and show that interactions occur between two SI 62D elements, but not between SI 62D and the gypsy insulator, limiting the scope of genomic gypsy insulator interactions. Enhancer blocking by SI 62D requires fewer Su(Hw)-binding sites than needed for gypsy insulator function, with these target regions having distinct zinc-finger requirements for in vivo Su(Hw) association. These observations led to an investigation of the role of the Su(Hw) zinc-finger domain in insulator function. Using a combination of in vitro and in vivo studies, we find that this domain makes sequence-dependent and -independent contributions to in vivo chromosome association, but is not essential for enhancer or silencer blocking. These studies extend our understanding of the properties of Su(Hw) and the endogenous genomic regions to which this protein localizes.
EUKARYOTIC genomes contain multiple classes of DNA elements that regulate transcription. One class includes insulators that restrict and define interactions between enhancers, silencers and promoters. Insulators have been identified on the basis of one of two properties (Kuhn and Geyer 2003; Capelson and Corces 2004; Recillas-Targa et al. 2004; Brasset and Vaury 2005; Valenzuela and Kamakaka 2006). First, insulators block enhancers when placed between an enhancer and a promoter (Geyer and Corces 1992; Kuhn and Geyer 2003). Enhancer blocking does not inactivate transcriptional regulatory elements, but prevents communication between enhancers and the target promoter (Cai and Levine 1995; Scott and Geyer 1995). Second, insulators act as barriers that protect the expression of transgenes from chromatin-silencing effects, including restricting the action of silencers and inhibiting the spread of heterochromatin (Roseman et al. 1995; Festenstein et al. 1996; Mallin et al. 1998; Pikaart et al. 1998; Yannaki et al. 2002; Jakobsson et al. 2004). Insulators are fundamental components of eukaryotic genomes that are involved in multiple processes, ranging from centromere organization in yeast to imprinting in mammals (Noma et al. 2001; Engel et al. 2006; Yoon et al. 2007).
The gypsy insulator is a well-characterized element in the Drosophila genome. This insulator resides in the 5′ untranslated region of the gypsy retrovirus and is responsible for mutations caused by insertion of this retrovirus into the regulatory regions of several genes (Modolell et al. 1983; Geyer et al. 1988; Peifer and Bender 1988; Spana et al. 1988). The gypsy insulator consists of 12 direct repeats of a YRYTGCATAYYY motif, where Y represents a pyrimidine and R represents a purine, separated by an AT-rich spacer. Direct tests have shown that the gypsy insulator blocks a wide variety of enhancers (Dorsett et al. 1989; Geyer and Corces 1992; Scott and Geyer 1995; Hagstrom et al. 1996; Cai and Levine 1997), protects against the repressive effects of a Polycomb response element (PRE; Sigrist and Pirrotta 1997; Mallin et al. 1998), and partially prevents heterochromatic silencing of transgenes inserted into centric regions (Roseman et al. 1993, 1995). These observations demonstrate that the gypsy insulator has both properties of insulators and shows a versatile capacity for defining regulatory interchanges.
Several proteins that are required for gypsy insulator function have been identified. An essential component is the Suppressor of Hairy-wing [Su(Hw)] protein that binds this insulator through a centrally located 12-zinc-finger domain (Parkhurst et al. 1988; Spana and Corces 1990; Harrison et al. 1993). Su(Hw) binding establishes a platform for protein–protein interactions that includes E(y)2/Sus1 (Kurshakova et al. 2007) and two BTB/POZ domain proteins, Modifier of mdg4 67.2 (Mod67.2) (Georgiev and Gerasimova 1989; Gerasimova et al. 1995) and centrosomal protein 190 (CP190) (Pai et al. 2004). Genetic studies indicate that Mod67.2 and CP190 are required for both enhancer and silencer-blocking effects of the gypsy insulator, while E(y)2/Sus1 is required only for barrier activity. These findings imply that proteins associated with the gypsy insulator make different contributions to the properties of this element.
The Su(Hw) protein associates with hundreds of non-gypsy regions within Drosophila euchromatin. Bioinformatic and biochemical approaches reveal that these endogenous regions have an extended consensus sequence of 20 bp relative to the 12-bp gypsy site, with plasticity in the TGCATA core (Parnell et al. 2006; Ramos et al. 2006; Adryan et al. 2007). The non-gypsy Su(Hw) regions do not contain clustered Su(Hw)-binding sites, with the vast majority carrying a single copy of the consensus sequence located in noncoding sequences. The absence of clustering was unexpected, as studies using synthetic insulators generated from a gypsy insulator Su(Hw)-binding site showed a requirement for at least four closely spaced Su(Hw)-binding sites for enhancer blocking (Scott et al. 1999). These observations have raised the question of whether endogenous sites are insulators. Studies of the enhancer-blocking properties of fragments containing endogenous Su(Hw) sites showed that these regions prevented enhancer–promoter communication, suggesting insulator activity, although the strength of the block was weaker than found for synthetic Su(Hw) and gypsy insulators (Golovnin et al. 2003; Parnell et al. 2003, 2006; Ramos et al. 2006). These observations are consistent with the previous findings that increasing the number of Su(Hw)-binding sites strengthens insulator activity (Hoover et al. 1992; Hagstrom et al. 1996).
We are studying endogenous Su(Hw) sites to determine whether these sequences have similar characteristics to those defined for the gypsy insulator. Here, we focused on an endogenous Su(Hw) cluster located at cytological position 62D in the polytene chromosomes, previously shown to have enhancer-blocking activity (Parnell et al. 2006). We demonstrate that this Su(Hw) insulator (SI) 62D displays a subset of gypsy insulator properties. Enhancer blocking by SI 62D requires fewer Su(Hw)-binding sites than needed for gypsy insulator function, with these target regions having distinct zinc fingers needed for in vivo association of the Su(Hw) protein. As models of insulator function predict that the gypsy insulator forms chromatin loop domains through interactions with endogenous Su(Hw) insulators (SIs) that limit the action of transcriptional control elements, we tested pairing interactions between SI 62D and the gypsy insulator, finding evidence that the scope of gypsy interactions do not extend to SI 62D. Finally, we addressed the role of the zinc-finger domain in insulator function, showing that this domain makes sequence-dependent and -independent contributions to in vivo chromosome association, but is not essential for enhancer or silencer blocking. These studies extend our understanding of the properties of endogenous Su(Hw)-binding regions and the function of the Su(Hw) protein.
MATERIALS AND METHODS
Genetic and phenotypic analyses:
Fly stocks were maintained at 25°, 70% humidity on standard corn meal and agar medium. Phenotypic analysis of yellow (y) gene expression depended on cuticle pigmentation analysis completed as described previously (Morris et al. 1999), using 3- to 4-day-old females. Here “wing” refers to the wing blade and “body” refers to pigmentation in the abdominal stripes. A score of 1 represents the null phenotype, a score of 2 represents pigmentation associated with flies carrying a gypsy insulator inserted into the endogenous yellow gene between the wing and body enhancer, a score of 3 represents intermediate pigmentation, and scores of 4–5 represent wild-type coloration. Scores were determined using a series of five parallel crosses. A plus sign indicates that the average level of pigmentation was slightly greater than that of the corresponding control. We consider differences in pigmentation significant only if the score differs by one or more units (Morris et al. 1999). Phenotypic analysis of white (w) gene expression depended on eye pigmentation analysis. Eye colors were determined on a color scale: white, pale yellow, yellow, orange, dark orange, brown, and red, representing increasing levels of transcription. At least three independent crosses were used in the phenotypic analyses.
Germline transformation was used to generate transgenic flies. P transposase was injected at a concentration of 400 μg/ml, with the “wings-clipped” helper plasmid pπ 25.7 at a concentration of 200 μg/ml into the host strain y1 w67c23. Southern analysis determined the number and integrity of the inserted transposons.
Enhancer-blocking transposons:
To generate transposons containing the SI 62D, we amplified a 426-bp region from cytological position 62D that included four predicted Su(Hw)-binding sites (Parnell et al. 2006). SI 62D was inserted into a NotI site at 900 relative to the yellow transcription start site, downstream of the wing and body enhancers. Insertions were selected for orientation, resulting in the generation of P[62D DNF] and P[62D DNR]. P[62D UP] was generated by cloning SI 62D into an intermediate vector containing flanking SalI sites and by subsequently inserting these sequences into an XhoI site at −2800 relative to the yellow transcription start site, upstream of the wing and body enhancers. P[62D 2DNR] was generated by cloning a blunt-ended fragment containing the four SI 62D Su(Hw) sites into the Eco47III site located at −900 relative to the yellow transcription start site and by screening for clones that contained multiple copies. P[62D Gyp-DNR] was generated by inserting a lox P flanked DNA fragment containing gypsy and SI 62D separated by 2 kb of λDNA into the Eco47III site. Previous studies used this 2-kb λDNA fragment in tests of insulator neutralization (Kuhn et al. 2003).
Chromatin immunoprecipitation:
Chromatin was prepared from third instar larvae as described in Parnell et al. (2003). Briefly, a nuclear suspension (∼109 nuclei/ml) was crosslinked with 1% formaldehyde at room temperature for 5 min. Nuclei were washed and lysed, and chromatin was sheared to an average length of ∼700 bp by sonication. In each chromatin immunoprecipitation (ChIP) experiment, a chromatin solution containing ∼20 μg of genomic DNA was incubated with either specific or nonspecific antibody. Immunoprecipitation and wash conditions were performed as described in (Parnell et al. 2003). Diluted input DNA (1:100) and precipitated ChIP DNA were used in PCR reactions. In each case, PCR reactions were set up and manually stopped at different consecutive cycle numbers. Products resulting from amplification cycles, usually between 21 and 25 cycles, were run on a polyacrylamide gel and visually detected using ethidium bromide, and the fluorescence signal was captured by digital photography for quantitation. Data were considered acceptable only when reactions showed linear amplification of the PCR products, such that the product in consecutive cycles increased approximately twofold (±0.3). To determine percentage of input (Figure 1), at least three separate immunoprecipitation experiments from at least two different chromatin preparations were analyzed using our manual PCR procedure. ChIP for Su(Hw) was performed using a rabbit anti-Su(Hw) antibody generously provided by Victor Corces. Normal rabbit IgG (Sigma) was used as a nonspecific antibody control. Antibody–chromatin complexes were captured with protein A Sepharose beads (Sigma). Primer sequences used in PCR for the ChIP analysis will be provided upon request.
Protein purification and electrophoretic mobility shift assay:
Full-length wild-type and mutant Su(Hw) proteins were expressed and purified from Escherichia coli DE3 cells. Two mutant proteins were studied; Su(Hw)f carries a cysteine-to-tyrosine substitution of amino acid (aa) 525, leading to inactivation of zinc finger 10, while Su(Hw)E8 carries a histidine-to-tyrosine substitution of aa 459, leading to inactivation of zinc finger 7. Each su(Hw) cDNA was cloned into a modified pET21a expression vector (Novagen) that contained an amino-terminal T7 tag and a carboxyl-terminal FLAG tag followed by a six His tag. Su(Hw) protein was induced by IPTG overnight at 18°. After harvesting, cells were lysed by sonication and the lysate was cleared by centrifugation at 45,000 rpm for 45 min. Partial purification of the Su(Hw) protein involved a two-step chromatography, first on Ni-NTA resin (QIAGEN) followed by salt elution from Mono Q (GE Healthcare Life Sciences), as described previously (Parnell et al. 2006).
Apparent DNA-binding affinities were determined using an electrophoretic mobility shift assay (EMSA). DNA fragments were isolated from TOPO TA clones by EcoRI digestion and end labeled using [32P]dATP and Klenow. For each reaction, 2 fmol of labeled DNA (∼1000–10,000 cpm) was incubated with 0, 0.003, 0.01, 0.03, 0.1, 0.3, and 1 μg of Su(Hw) protein in 1× binding buffer (15 mm HEPES, pH 7.6, 100 mm KCl, 250 μm ZnCl2, 10% glycerol). After a 15-min incubation at room temperature, reaction products were separated by electrophoresis overnight on 1% agarose 0.25× TBE gels at 4°. Dried gels were analyzed by autoradiography and counts in bound and unbound bands were measured using an Instant Imager (Packard). Affinity constants were determined by nonlinear least-square analysis of a Langmuir binding equation for noncooperative binding using Kaleidagraph (Synergy Software).
LexA-tethering system:
Expressor lines were generated using P transposons that encoded the Su(Hw) protein carrying a wild-type or mutant zinc-finger DNA-binding domain fused in frame to the 202-aa DNA-binding domain (DBD) of the E. coli LexA protein. cDNA sequences corresponding to each fusion protein were cloned downstream of the Actin 5C promoter that is active throughout most of development (Fyrberg et al. 1983). FL-LexA represents a fusion of the full-length Su(Hw) protein (941 aa) with the LexA DBD, ΔZnF-LexA represents a fusion protein with a mutant Su(Hw) deleted for the entire zinc-finger domain (aa 220–620), and mF10-LexA represents a fusion protein carrying the cysteine-to-tyrosine substitution at aa 525, mimicking the protein encoded by the su(Hw)f allele (Harrison et al. 1993).
Two transposons were used to assess the activity of the Su(Hw)-LexA fusion proteins, each carrying the mini-yellow and mini-white genes as reporters (Mallin et al. 1998). In these transposons, a LexA-binding region (BR) that contained six LexA-binding sites was inserted between the enhancer or silencer and the promoter of the reporter transgene. The enhancer-blocking reporter, P[Y L En L w], carried two LexA BRs, one inserted between the wing and body enhancers and the yellow promoter and one inserted between the white eye enhancer and the white promoter. Downstream of the white gene, a gypsy insulator was inserted to protect against position effects (Roseman et al. 1993). The silencer-blocking reporter, P[y PRE L w], contained a LexA BR inserted between the 1.6-kb PRE from the Ubx locus (Mallin et al. 1998) and the enhancer of the mini-white gene. Transgenic flies carrying an expressor or a reporter transposon were mated at 25°. Progeny from these crosses were isolated and aged for 2–3 days for phenotypic analyses.
RESULTS
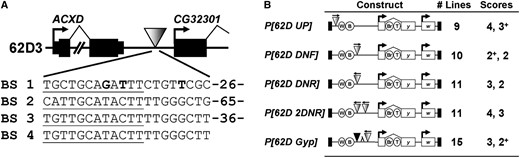
SI 62D contains a cluster of four predicted Su(Hw)-binding sites located in the intergenic region that separates the ACXD and CG32301 genes (Figure 1). Previous studies demonstrated that the Su(Hw) protein binds this region in vivo (Parnell et al. 2006). To examine the insulator properties of SI 62D, several P transposons that carried the yellow cuticle pigmentation gene were generated (Parnell et al. 2006) (Figure 1). This gene serves as an excellent reporter, as expression depends on four independent enhancers, located upstream (wing and body) and downstream (bristle and tarsal claw) of the transcription start site (Chia et al. 1986; Geyer and Corces 1987), thereby discriminating between insertion of a global repressor or a position-dependent enhancer blocker. SI 62D was inserted into the yellow regulatory regions upstream (P[62D UP]) and downstream (P[62D DNF] and P[62D DNR]) of the wing and body enhancers. Multiple transgenic flies carrying single transposon insertions were generated and examined for cuticle color in adults. These studies showed that P[62D UP] flies had dark pigmentation of all cuticle structures (wing and body scores of 4 and 3+), while P[62D DNF] and P[62D DNR] flies had light pigmentation in the wing and body (scores of 2+/3 and 2; Figure 1) and dark bristle pigmentation (data not shown). Flies carrying the P[62D DNF] and P[62D DNR] transposons had similar pigmentation scores to those carrying a reporter transposon with the gypsy insulator inserted between the yellow wing and body enhancers and promoter (scores 2+ and 2) (Parnell et al. 2006). These results demonstrate that SI 62D provides a strong, position-dependent, orientation-independent block of enhancer activated transcription.
Properties of SI 62D. (A) Map of cytological location 62D showing the position of the SI 62D insulator (inverted shaded triangle). SI 62D is located 424 nt downstream of ACXD and 188 nt upstream of CG32301. Genes are shown as solid rectangles, with the direction of transcription shown by the bent arrows. The DNA sequence of the four predicted Su(Hw)-binding sites is shown, with the number of base pairs separating each site indicated. The nucleotide differences between the 62D BS 1 and the Su(Hw)-binding site consensus sequence (Adryan et al. 2007) are shown in boldface type. The 12 nucleotides matching the gypsy insulator consensus are underlined. (B) Structure of transposons used to define the properties of SI 62D. The reporter yellow (y) gene contains four enhancers: wing (W), body (B), bristle (Br), and tarsal claws (T), with the white (w) gene used as the transformation marker. Reporter genes are cloned into a P transposon (P ends shown as solid boxes). SI 62D is shown as a shaded triangle and the gypsy insulator as a solid triangle. The relative orientation of SI 62D is shown by the horizontal arrow. “λ” indicates the spacer DNA isolated from the λ-bacteriophage. The middle column lists the number of transgenic lines studied, while the right column summarizes the average pigmentation score obtained for the wing and body cuticle.
Insulator neutralization studies of SI 62D:
The gypsy insulator displays pairing interactions such that when two insulators are placed between an enhancer and a promoter, enhancer blocking is lost. These effects, known as insulator neutralization, are a property of some, but not all, insulators (Cai and Shen 2001; Muravyova et al. 2001; Kuhn et al. 2003; Majumder and Cai 2003; Gruzdeva et al. 2005; Chetverina et al. 2008). We tested whether the SI 62D insulator displayed insulator neutralization through studies of transgenic flies carrying the P[62D 2DNR] transposon that carried a yellow gene with two SI 62D insulators inserted between the wing and body enhancers and promoter (Figure 1). Analysis of cuticle color in P[62D 2DNR] adult flies showed dark pigmentation of the wing and body (scores of 4 and 3), indicating a loss of enhancer blocking and insulator neutralization. To determine whether pairing interactions of SI 62D extended to the gypsy insulator, we generated P[62D Gyp-DNR] flies that carried a transposon with a yellow gene that had SI 62D and the gypsy insulator inserted between the wing and body enhancer and promoter. Surprisingly, we found that P[62D Gyp-DNR] flies showed an enhancer block, with adult flies having light pigmentation in the wing and body (3 and 2+; Figure 1), a level of cuticle color that was similar to flies carrying the single insulator transposons. Although the spacing of insulators differs in P[62D 2DNR] and P[62D Gyp-DNR], we do not feel that this difference accounts for the absence of neutralization for the following reasons. First, the separation distance in the P[62D Gyp-DNR] transposon reproduced previous spacing used to demonstrate neutralization between gypsy insulators (Kuhn et al. 2003). Second, pairing interactions between insulator does not appear to be sensitive to spacing, as interactions are observed in situations where insulators are separated between 50 bp and 5 kb (Cai and Shen 2001; Muravyova et al. 2001). Our observations imply that the pairing interactions of SI 62D are restricted and cannot occur with the gypsy insulator.
The su(Hw)f allele affects endogenous insulators differently from the gypsy insulator:
Mutations in the su(Hw) gene reverse the enhancer-blocking effects of the gypsy insulator. A commonly used su(Hw) mutant background is su(Hw)v/su(Hw)f. The su(Hw)v allele carries a small deletion that encompasses the su(Hw) promoter and does not encode a protein. The su(Hw)f allele carries a base-pair substitution that results in a cysteine-to-tyrosine substitution within zinc finger 10, inactivating this zinc finger (Harrison et al. 1992, 1993). On the basis of studies of the gypsy insulator, we predicted that enhancer blocking would be lost in P[62D DNR], su(Hw)v/su(Hw)f flies. However, we found that P[62D DNR], su(Hw)v/su(Hw)f mutant flies had light wing and body pigmentation (scores of 3 and 2+; data not shown), indicating that enhancer blocking was retained. These data imply that the Su(Hw)f protein binds SI 62D in vivo.
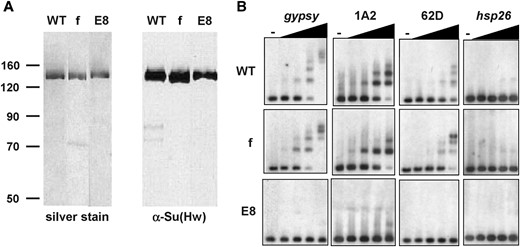
ChIP was used to test whether Su(Hw)f retained the ability to associate with SI 62D (Figure 2, Table 1). Chromatin was isolated from wild-type and su(Hw)v/su(Hw)f third instar larvae and immunoprecipitated using either a Su(Hw) or a nonspecific antibody. DNA enrichment was analyzed by PCR amplification using primers that flanked SI 62D, the gypsy and the 1A-2 insulator (positive controls), and the hsp26 coding region (negative control). In a wild-type su(Hw) background, PCR amplification products were obtained for the gypsy insulator, 1A-2, and SI 62D (Figure 2A). In the su(Hw)v/su(Hw)f mutant background, amplification of the gypsy and 1A-2 insulators was lost, whereas a product was obtained from PCR using SI 62D primers (Figure 2A). These analyses suggest that Su(Hw) binding to SI 62D did not decrease in the su(Hw)v/su(Hw)f mutant background. We tested whether the previously identified endogenous SIs (Parnell et al. 2006) bound the Su(Hw)f protein in vivo and found that 50A and 87E were PCR amplified in Su(Hw)-enriched chromatin (Figure 2B). These data indicate that the loss of in vivo Su(Hw) binding by mutation of zinc finger 10 depends on the genomic context (Figure 2B, Table 1).
ChIP analysis of endogenous Su(Hw) sites in a wild-type and su(Hw) mutant background. (A) PCR analysis. Shown are representative examples of PCR products obtained from chromatin material that was either directly purified (input, or IN) or immunoprecipitated with either Su(Hw) antibody (SH) or a nonspecific rabbit (NS) antibody. Amplified regions correspond to the gypsy insulator, hsp26 coding region, and Su(Hw) insulators 1A-2 and 62D. Chromatin was derived from wild-type [Su(Hw)+/+] and mutant [Su(Hw)v/f] larvae. (B) ChIP analysis of endogenous SIs. The percentage of input was determined by quantifying the intensity of the PCR product in the immunoprecipitated fraction relative to input. Wild-type ratios are indicated by solid bars, while mutant ratios are indicated by shaded bars. The average of at least three ChIP experiments is shown, with error bars indicating the standard deviation.
Summary of DNA-binding properties of wild-type and mutant Su(Hw) proteins
. | . | . | Apparent Ka (m−1)a . | ChIPb . | |||
|---|---|---|---|---|---|---|---|
| Name . | No. of Su(Hw) sites . | Spacing . | Su(Hw)WT . | Su(Hw)f . | Su(Hw)E8 . | su(Hw)+ . | su(Hw)f . |
| gypsy | 12 | 14–23 | 9.7 × 107 | 8.2 × 107 | ≤3 × 106 | +++ | − |
| hsp26 | 0 | — | ≤2.0 × 106 | ≤2 × 106 | ≤1 × 106 | − | − |
| 1A-2 | 2 | 37 | 6.6 × 107 | 6.8 × 107 | ≤1 × 106 | ++ | − |
| 62D | 4 | 34, 73, 44 | 4.3 ×107 | 7.7 × 107 | ≤1 × 106 | +++ | +++ |
. | . | . | Apparent Ka (m−1)a . | ChIPb . | |||
|---|---|---|---|---|---|---|---|
| Name . | No. of Su(Hw) sites . | Spacing . | Su(Hw)WT . | Su(Hw)f . | Su(Hw)E8 . | su(Hw)+ . | su(Hw)f . |
| gypsy | 12 | 14–23 | 9.7 × 107 | 8.2 × 107 | ≤3 × 106 | +++ | − |
| hsp26 | 0 | — | ≤2.0 × 106 | ≤2 × 106 | ≤1 × 106 | − | − |
| 1A-2 | 2 | 37 | 6.6 × 107 | 6.8 × 107 | ≤1 × 106 | ++ | − |
| 62D | 4 | 34, 73, 44 | 4.3 ×107 | 7.7 × 107 | ≤1 × 106 | +++ | +++ |
Ka, apparent association constant, where m−1 refers to reverse molar.
ChIP results in the indicated su(Hw) mutant background with the degree of in vivo Su(Hw) association represented by no (−) to strong association (+++).
Summary of DNA-binding properties of wild-type and mutant Su(Hw) proteins
. | . | . | Apparent Ka (m−1)a . | ChIPb . | |||
|---|---|---|---|---|---|---|---|
| Name . | No. of Su(Hw) sites . | Spacing . | Su(Hw)WT . | Su(Hw)f . | Su(Hw)E8 . | su(Hw)+ . | su(Hw)f . |
| gypsy | 12 | 14–23 | 9.7 × 107 | 8.2 × 107 | ≤3 × 106 | +++ | − |
| hsp26 | 0 | — | ≤2.0 × 106 | ≤2 × 106 | ≤1 × 106 | − | − |
| 1A-2 | 2 | 37 | 6.6 × 107 | 6.8 × 107 | ≤1 × 106 | ++ | − |
| 62D | 4 | 34, 73, 44 | 4.3 ×107 | 7.7 × 107 | ≤1 × 106 | +++ | +++ |
. | . | . | Apparent Ka (m−1)a . | ChIPb . | |||
|---|---|---|---|---|---|---|---|
| Name . | No. of Su(Hw) sites . | Spacing . | Su(Hw)WT . | Su(Hw)f . | Su(Hw)E8 . | su(Hw)+ . | su(Hw)f . |
| gypsy | 12 | 14–23 | 9.7 × 107 | 8.2 × 107 | ≤3 × 106 | +++ | − |
| hsp26 | 0 | — | ≤2.0 × 106 | ≤2 × 106 | ≤1 × 106 | − | − |
| 1A-2 | 2 | 37 | 6.6 × 107 | 6.8 × 107 | ≤1 × 106 | ++ | − |
| 62D | 4 | 34, 73, 44 | 4.3 ×107 | 7.7 × 107 | ≤1 × 106 | +++ | +++ |
Ka, apparent association constant, where m−1 refers to reverse molar.
ChIP results in the indicated su(Hw) mutant background with the degree of in vivo Su(Hw) association represented by no (−) to strong association (+++).
The Su(Hw) protein has a large zinc-finger domain of 12 fingers (Parkhurst et al. 1988), reminiscent of the zinc-finger domain of the vertebrate CTCF insulator protein (Lobanenkov et al. 1990). Studies of CTCF have shown that distinct combinations of the 11 zinc fingers are used to bind different genes (Filippova et al. 1996). On the basis of these findings, we postulated that endogenous Su(Hw)-binding regions might require different zinc fingers for DNA recognition and binding than needed for the gypsy insulator, explaining the observation that the loss of zinc finger 10 did not alter in vivo association of Su(Hw) to SI 62D and other endogenous sites. However, inspection of the SI 62D DNA sequence did not provide insights into how this discrimination might be achieved, as the Su(Hw) sites in SI 62D match both the gypsy and the endogenous consensus sequence (Figure 1) (Parnell et al. 2006; Adryan et al. 2007).
We tested the in vitro binding properties of the Su(Hw)f protein to determine whether the loss of zinc finger 10 disrupted DNA recognition to a subset of SIs and the gypsy insulator. For these studies, wild-type and mutant Su(Hw) proteins were expressed in E. coli and purified (Figure 3A). EMSAs were performed with radiolabeled probes corresponding to 1A-2, SI 62D, the gypsy insulator, and a fragment containing the hsp26 coding region as a negative control (Figure 3B). As shown previously (Parnell et al. 2006), the wild-type Su(Hw) protein produced five to six distinct migrating species when incubated with the gypsy insulator, indicating that multiple Su(Hw)-binding sites can be occupied simultaneously. However, we did not observe 12 distinctly migrating species, as might be predicted if all binding sites were occupied. Two explanations are possible for these observations. First, the Su(Hw) protein may not be able to bind to all 12 sites simultaneously due to physical occlusion of neighboring sites. Alternatively, our gel system may not resolve DNA fragments that contain more than six bound proteins. Su(Hw) binding to SI 62D was tested using the wild-type protein. We found that the wild-type Su(Hw) protein produced three shifted species (Figure 3B), suggesting that one binding site of SI 62D site might not be recognized. We note that even though the sequence of binding site 1 closely matches the endogenous consensus, three of the strongly preferred nucleotides (8, 10, and 17) are absent (Figure 1). These findings predict that these three nucleotides play a key role in Su(Hw) recognition of the consensus site.
Su(Hw)f associates with the gypsy insulator, 1A-2, and 62D in vitro. (A) Analysis of purified Su(Hw) protein. Wild-type (WT) and mutant Su(Hw) proteins were purified from E. coli, run on an SDS–polyacrylamide gel, and detected by silver staining (left) or with a Su(Hw) antibody (right). Su(Hw)f (f) carries a point mutation that inactivates zinc finger 10, while Su(Hw)E8 (E8) carries a point mutation that inactivates zinc finger 7. The positions of the protein size markers (in kilodaltons) are shown at the left. (B) EMSA analyses. Results are shown for 32P-labeled DNAs corresponding to four DNA fragments (the gypsy, 1A-2, and 62D insulators and the hsp26 coding region) that were incubated with no (-) or increasing amounts of the wild-type (WT) or Su(Hw) mutant proteins (f, E8). The amount of recombinant protein was increased threefold in each lane, beginning at 0.003 μg.
The DNA-binding parameters for two Su(Hw) mutant proteins, Su(Hw)E8 and Su(Hw)f, were determined (Figure 3B, Table 1). The Su(Hw)E8 protein that has an inactive zinc finger 7 was previously shown not to bind DNA in vitro (Harrison et al. 1993; Kim et al. 1996). As expected, our studies showed that this protein did not produce slower migrating species with any DNA tested (Figure 3). In contrast, we found that Su(Hw)f bound DNA fragments that contained Su(Hw) sites, but did not bind fragments that lacked these sequences (hsp26 coding DNA). Interestingly, the apparent binding affinities associated with Su(Hw)f binding were similar to those determined for the wild-type protein (Table 1). Taken together, these data suggest that the inability of the Su(Hw)f protein to localize in vivo to the gypsy insulator and some endogenous SIs does not reflect a loss of DNA recognition, as previously suspected (Kim et al. 1996). Instead, these data demonstrate a second role for the Su(Hw) zinc-finger domain in chromosome association. As zinc fingers are versatile protein domains that direct DNA, RNA, and protein association (Gamsjaeger et al. 2007), we postulate that the context-dependent difference in the requirement for zinc finger 10 may reflect the need for interaction with a protein(s) that modifies chromatin to facilitate recognition of DNA.
Use of a LexA-tethering system to test the role of the Su(Hw) zinc-finger domain in insulator activity:
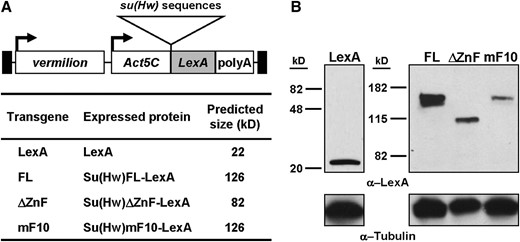
We wondered whether the Su(Hw) zinc-finger domain contributes to insulator activity outside of the role in in vivo chromosome association. We reasoned that if the need for the DNA binding of the zinc-finger domain were circumvented, then we could test the role of this domain in enhancer and silencer blocking. To this end, we developed a Su(Hw)-LexA tethering system. In this system, expressor lines were established wherein Su(Hw)-LexA DBD proteins were generated using the Actin5C promoter (Figure 4). Four expressor lines were generated, including LexA alone (LexA), the full-length Su(Hw) fused to LexA (FL-LexA), a mutant Su(Hw) deleted for the entire DNA-binding domain fused to LexA (ΔZnF-LexA), and a mutant Su(Hw) lacking zinc finger 10 fused to LexA (mF10-LexA). Western analyses of proteins extracted from transgenic flies carrying these expressor transposons were conducted to determine whether proteins of the appropriate size accumulated. Using an antibody directed against the LexA protein, we found that each expressor line produced a protein of the appropriate molecular weight, with similar levels of accumulation of LexA, ΔZnF-LexA, and mF10-LexA and greater accumulation of the FL-LexA protein (Figure 4).
Expressor transgenes used in LexA-tethering studies. (A, top) General structure of the expressor transgenes. Expressor transposons carried the vermilion gene for use as an injection marker. The Actin 5C promoter (Act5C) drives expression of a fusion gene carrying su(Hw) cDNA sequences (triangle), the LexA coding sequence (LexA shaded box), and the α-tubulin polyadenylation sequence (polyA). Arrows indicate transcription start sites. Solid boxes indicate P-element ends. (Bottom) Table of expressor transgenes. Predicted sizes for each LexA protein were calculated from amino acid sequences of entire proteins, including LexA DNA-binding domain only (LexA); full-length Su(Hw)-LexA (FL); ΔZnF-LexA, deleted for the zinc-finger domain (ΔZnF); and mF10-LexA (mF10), mutant for zinc finger 10. (B) Western analysis of LexA fusion proteins extracted from expressor lines. Total protein was extracted from 1- to 2-day-old adult flies, and one fly equivalent was loaded in each lane. Fusion proteins were visualized with an anti-LexA antibody (1:1000). α-Tubulin was used as a loading control (1:400,000). Migration of protein size markers (in kilodaltons) is shown.
Two reporter transposons were used in the tethering assay, each carrying a LexA BR containing six sites inserted between the regulatory elements to be tested and the promoter for the yellow or white reporter genes (Figure 5). The enhancer-blocking reporter (P[y L En L w]) contained two LexA BRs, one inserted between the yellow enhancers and promoter and one between the white eye enhancer and promoter. The silencer-blocking reporter (P[y PRE L w]) contained a single LexA BR inserted between the Ubx PRE and the white promoter. This PRE directs Polycomb-dependent silencing of nearby reporter genes, including yellow and white, when present in a transposon (Sigrist and Pirrotta 1997; Mallin et al. 1998; Comet et al. 2006). Gene silencing depends upon the genomic site of insertion, with most, but not all, positions conferring repression. Multiple independent insertion lines for each reporter transposon were obtained and lines with single transposon insertions were studied.
Summary of results obtained in the LexA-tethering assay. (Left) Percentage of lines that reconstituted enhancer blocking (top) and silencing blocking (bottom) using the yellow gene as a reporter. The numbers in parentheses represent the lines showing insulator activity relative to the total number tested. (Middle) Structures of the two reporter transposons. P[y L En L w] contains a LexA BR (L in shaded box) of six repeated sites inserted between the yellow wing and body enhancers (ovals marked W and B) and yellow (y) promoter and between the eye enhancer (oval marked E) and white promoter (w). A gypsy insulator (inverted solid triangle) is located at the 3′-end of the white gene to prevent position effects. P[y PRE L w] contains the LexA BR (L in shaded boxes) inserted between the white eye enhancer and the PRE. Solid boxes indicate P-element ends. (Right) Percentage of lines that reconstituted enhancer blocking (top) and silencing blocking (bottom) using the white gene as a reporter.
Enhancer blocking can be reconstituted by a tethered Su(Hw) protein lacking the zinc-finger domain:
Transgenic flies carrying the enhancer-blocking reporter P[y L En L w] had dark cuticle and eye pigmentation, indicating enhancer-activated transcription of the yellow and white reporter genes. Flies representing each independent P[y L En L w] reporter line were crossed with expressor flies and phenotypes of the resulting adult progeny were examined to determine whether the encoded LexA protein had blocking activity (Figure 5, Tables 2 and 3). We reasoned that if enhancer blocking were established at the LexA BRs, then lighter cuticle and eye coloration would be observed. Reporter flies expressing LexA showed no change in phenotype, demonstrating that LexA alone does not disrupt enhancer–promoter communication. In contrast, reporter flies expressing FL-LexA protein showed light cuticle pigmentation in progeny obtained from two of five lines tested, and light eye color in four of five lines tested (Tables 2 and 3). Curiously, in three reporter lines, enhancer blocking was confined to one of the two reporter genes in the transposon. These results might reflect that amounts of the fusion protein are limited such that the two LexA BRs compete for binding of the fusion protein. In one case, we found that the cross between the FL expressor and the P[y L En L w] reporter generated progeny that had light eye and cuticle pigmentation, suggesting that robust blocking of both sets of enhancers is possible. Taken together, our data indicate that full-length Su(Hw) protein directed to chromosomes using the LexA DBD reconstitutes enhancer blocking at certain genomic locations.
Pigmentation scores (wing, body) associated with yellow expression in the presence of the indicated LexA protein
. | LexA expressor line . | |||||
|---|---|---|---|---|---|---|
| Reporter line . | Line no. . | None . | LexA . | FL . | ΔZnF . | mF10 . |
| P[y L En L w] | 12 | 5, 5 | 5, 5 | 5, 4+ | 5, 4+ | 5, 4+ |
| 24 | 5, 5 | 5, 5 | 5, 4+ | 5, 4+ | 5, 4+ | |
| 32 | 5, 5 | 5, 5 | 5, 4+ | 5, 4+ | 5, 4+ | |
| 36-1 | 5, 5 | 5, 5 | 2, 2 | 4, 4 | 4, 4 | |
| 36-2 | 5, 5 | 5, 5 | 2, 2 | 3, 3 | 3, 3 | |
| P[y PRE L w] | 21 | 4, 4+ | 4, 4+ | 4, 4+ | 4+, 4+ | 4, 4+ |
| 26 | 3, 3+ | 3, 3+ | 4, 4+ | 3+, 3+ | 3, 3+ | |
| 64 | 1, 1 | 1, 1 | 2, 2 | 1, 1 | 1, 1 | |
| 66 | 2, 3 | 2, 3 | 5, 5 | 4, 4+ | 4, 4+ | |
| 104 | 3, 3 | 3, 3 | 4, 4+ | 4, 4+ | 4, 4 | |
. | LexA expressor line . | |||||
|---|---|---|---|---|---|---|
| Reporter line . | Line no. . | None . | LexA . | FL . | ΔZnF . | mF10 . |
| P[y L En L w] | 12 | 5, 5 | 5, 5 | 5, 4+ | 5, 4+ | 5, 4+ |
| 24 | 5, 5 | 5, 5 | 5, 4+ | 5, 4+ | 5, 4+ | |
| 32 | 5, 5 | 5, 5 | 5, 4+ | 5, 4+ | 5, 4+ | |
| 36-1 | 5, 5 | 5, 5 | 2, 2 | 4, 4 | 4, 4 | |
| 36-2 | 5, 5 | 5, 5 | 2, 2 | 3, 3 | 3, 3 | |
| P[y PRE L w] | 21 | 4, 4+ | 4, 4+ | 4, 4+ | 4+, 4+ | 4, 4+ |
| 26 | 3, 3+ | 3, 3+ | 4, 4+ | 3+, 3+ | 3, 3+ | |
| 64 | 1, 1 | 1, 1 | 2, 2 | 1, 1 | 1, 1 | |
| 66 | 2, 3 | 2, 3 | 5, 5 | 4, 4+ | 4, 4+ | |
| 104 | 3, 3 | 3, 3 | 4, 4+ | 4, 4+ | 4, 4 | |
Italic type indicates data that reflect phenotypic change due to insulator activity.
Pigmentation scores (wing, body) associated with yellow expression in the presence of the indicated LexA protein
. | LexA expressor line . | |||||
|---|---|---|---|---|---|---|
| Reporter line . | Line no. . | None . | LexA . | FL . | ΔZnF . | mF10 . |
| P[y L En L w] | 12 | 5, 5 | 5, 5 | 5, 4+ | 5, 4+ | 5, 4+ |
| 24 | 5, 5 | 5, 5 | 5, 4+ | 5, 4+ | 5, 4+ | |
| 32 | 5, 5 | 5, 5 | 5, 4+ | 5, 4+ | 5, 4+ | |
| 36-1 | 5, 5 | 5, 5 | 2, 2 | 4, 4 | 4, 4 | |
| 36-2 | 5, 5 | 5, 5 | 2, 2 | 3, 3 | 3, 3 | |
| P[y PRE L w] | 21 | 4, 4+ | 4, 4+ | 4, 4+ | 4+, 4+ | 4, 4+ |
| 26 | 3, 3+ | 3, 3+ | 4, 4+ | 3+, 3+ | 3, 3+ | |
| 64 | 1, 1 | 1, 1 | 2, 2 | 1, 1 | 1, 1 | |
| 66 | 2, 3 | 2, 3 | 5, 5 | 4, 4+ | 4, 4+ | |
| 104 | 3, 3 | 3, 3 | 4, 4+ | 4, 4+ | 4, 4 | |
. | LexA expressor line . | |||||
|---|---|---|---|---|---|---|
| Reporter line . | Line no. . | None . | LexA . | FL . | ΔZnF . | mF10 . |
| P[y L En L w] | 12 | 5, 5 | 5, 5 | 5, 4+ | 5, 4+ | 5, 4+ |
| 24 | 5, 5 | 5, 5 | 5, 4+ | 5, 4+ | 5, 4+ | |
| 32 | 5, 5 | 5, 5 | 5, 4+ | 5, 4+ | 5, 4+ | |
| 36-1 | 5, 5 | 5, 5 | 2, 2 | 4, 4 | 4, 4 | |
| 36-2 | 5, 5 | 5, 5 | 2, 2 | 3, 3 | 3, 3 | |
| P[y PRE L w] | 21 | 4, 4+ | 4, 4+ | 4, 4+ | 4+, 4+ | 4, 4+ |
| 26 | 3, 3+ | 3, 3+ | 4, 4+ | 3+, 3+ | 3, 3+ | |
| 64 | 1, 1 | 1, 1 | 2, 2 | 1, 1 | 1, 1 | |
| 66 | 2, 3 | 2, 3 | 5, 5 | 4, 4+ | 4, 4+ | |
| 104 | 3, 3 | 3, 3 | 4, 4+ | 4, 4+ | 4, 4 | |
Italic type indicates data that reflect phenotypic change due to insulator activity.
Eye color associated with white expression in the presence of the indicated LexA protein
. | Expressor line . | |||||
|---|---|---|---|---|---|---|
| Reporter line . | Line no. . | None . | LexA . | FL . | ΔZnF . | mF10 . |
| P[y L En L w] | 12 | Red | Red | Orange | Dark orange | Dark orange |
| 24 | Red | Red | Dark orange | Brown | Brown | |
| 32 | Red | Red | Light orange | Dark orange | Dark orange | |
| 36-1 | Red | Red | Dark orange | Brown | Brown | |
| 36-2 | Red | Red | Red | Red | Red | |
| P[y PRE L w] | 21 | Light orange variegated | Light orange variegated | Red | Red | Red |
| 26 | Orange variegated | Orange variegated | Orange | Brown | Brown | |
| 64 | Orange variegated | Orange variegated | Brown | Red | Brown | |
| 66 | Yellow | Yellow | Orange | Red | Red | |
| 104 | White | White | Light orange | Red | Brown | |
. | Expressor line . | |||||
|---|---|---|---|---|---|---|
| Reporter line . | Line no. . | None . | LexA . | FL . | ΔZnF . | mF10 . |
| P[y L En L w] | 12 | Red | Red | Orange | Dark orange | Dark orange |
| 24 | Red | Red | Dark orange | Brown | Brown | |
| 32 | Red | Red | Light orange | Dark orange | Dark orange | |
| 36-1 | Red | Red | Dark orange | Brown | Brown | |
| 36-2 | Red | Red | Red | Red | Red | |
| P[y PRE L w] | 21 | Light orange variegated | Light orange variegated | Red | Red | Red |
| 26 | Orange variegated | Orange variegated | Orange | Brown | Brown | |
| 64 | Orange variegated | Orange variegated | Brown | Red | Brown | |
| 66 | Yellow | Yellow | Orange | Red | Red | |
| 104 | White | White | Light orange | Red | Brown | |
Italic type indicates data that reflect phenotypic change due to insulator activity. The level of white expression is correlated with eye color in the descending scale: (high) red > brown > dark orange > orange > light orange > yellow > white (none).
Eye color associated with white expression in the presence of the indicated LexA protein
. | Expressor line . | |||||
|---|---|---|---|---|---|---|
| Reporter line . | Line no. . | None . | LexA . | FL . | ΔZnF . | mF10 . |
| P[y L En L w] | 12 | Red | Red | Orange | Dark orange | Dark orange |
| 24 | Red | Red | Dark orange | Brown | Brown | |
| 32 | Red | Red | Light orange | Dark orange | Dark orange | |
| 36-1 | Red | Red | Dark orange | Brown | Brown | |
| 36-2 | Red | Red | Red | Red | Red | |
| P[y PRE L w] | 21 | Light orange variegated | Light orange variegated | Red | Red | Red |
| 26 | Orange variegated | Orange variegated | Orange | Brown | Brown | |
| 64 | Orange variegated | Orange variegated | Brown | Red | Brown | |
| 66 | Yellow | Yellow | Orange | Red | Red | |
| 104 | White | White | Light orange | Red | Brown | |
. | Expressor line . | |||||
|---|---|---|---|---|---|---|
| Reporter line . | Line no. . | None . | LexA . | FL . | ΔZnF . | mF10 . |
| P[y L En L w] | 12 | Red | Red | Orange | Dark orange | Dark orange |
| 24 | Red | Red | Dark orange | Brown | Brown | |
| 32 | Red | Red | Light orange | Dark orange | Dark orange | |
| 36-1 | Red | Red | Dark orange | Brown | Brown | |
| 36-2 | Red | Red | Red | Red | Red | |
| P[y PRE L w] | 21 | Light orange variegated | Light orange variegated | Red | Red | Red |
| 26 | Orange variegated | Orange variegated | Orange | Brown | Brown | |
| 64 | Orange variegated | Orange variegated | Brown | Red | Brown | |
| 66 | Yellow | Yellow | Orange | Red | Red | |
| 104 | White | White | Light orange | Red | Brown | |
Italic type indicates data that reflect phenotypic change due to insulator activity. The level of white expression is correlated with eye color in the descending scale: (high) red > brown > dark orange > orange > light orange > yellow > white (none).
We tested effects of tethering mutant Su(Hw) fusion proteins, carrying either a disrupted (mF10-LexA) or an absent (ΔZnF-LexA) zinc-finger domain (Figure 5, Tables 2 and 3). Flies carrying each of Su(Hw)-LexA zinc-finger expressor transposons were crossed to flies from each of the P[y L En L w] reporter lines. Similar to our results with the FL Su(Hw) protein, we found that the progeny from two of five lines had reduced cuticle pigmentation, while progeny from four of five lines tested had a light eye color (Tables 2 and 3). Overall, the effects of tethering ΔZnF-LexA or mF10-LexA were similar but slightly weaker than seen for tethering the FL-LexA protein (Figure 5). We attribute these weaker effects to the lower levels of accumulation of the mutant proteins (Figure 4). The fact that enhancer blocking was reconstituted in the absence of the Su(Hw) zinc-finger domain suggests that domains outside of the zinc-finger domain are sufficient for disruption of enhancer–promoter communication.
Silencer blocking can be established by a tethered Su(Hw) protein lacking the zinc-finger domain:
Transgenic flies carrying the P[y PRE L w] displayed phenotypes consistent with the presence of a silencer element within the transposon. Depending upon the genomic insertion site, the eye phenotypes of these flies ranged in color from white to a variegated orange, whereas levels of cuticle coloration were reduced relative to wild-type flies in four of five lines (Tables 2 and 3). Interestingly, the degree of white and yellow repression at a given genomic location did not always correlate. Previous studies demonstrated that PRE silencing is blocked by the insertion of a gypsy insulator between the PRE and target promoter (Sigrist and Pirrotta 1997; Mallin et al. 1998). We reasoned that if tethered Su(Hw)-LexA fusion proteins reconstituted barrier activity, then white and yellow gene expression would increase, as the PRE silencer would be blocked. Crosses between P[y PRE L w] reporter flies and flies carrying the LexA expressor transposon produced progeny that were indistinguishable from those lacking the expressor transposon, indicating that LexA alone has no capacity for silencer blocking. In contrast, crosses of the same reporter lines with the FL-LexA expressor transposon generated progeny with a darker eye color than was seen in flies that carried only the reporter and with a darker cuticle color in four of the five lines tested. These data demonstrate that tethering the full-length Su(Hw) protein between the PRE and white promoter prevents silencing effects conferred on both reporter genes.
Flies carrying the ΔZnF-LexA or mF10-LexA expressor transposons were crossed to flies from each of the P[y PRE L w] reporter lines and the phenotype of the progeny was determined. We found that white gene expression was increased in all lines, with levels of eye color that were comparable or increased relative to those found for tethering the FL-LexA protein. These data suggest that the ΔZnF-LexA and mF10-LexA fusion proteins provide a strong block. Examination of cuticle pigmentation suggested that tethering of ΔZnF-LexA or mF10-LexA improved yellow gene expression, even though the LexA BR was not between the PRE and yellow promoter. Such effects imply that silencing of the yellow gene depends on interactions between the transposon PRE and PRE sequences in the genome that lie upstream of the white gene (Sigrist and Pirrotta 1997). Taken together, we conclude that barrier activity of the Su(Hw) protein does not require the zinc-finger domain, when the protein is localized to the chromosome using an independent DNA-binding domain.
DISCUSSION
Endogenous Su(Hw)-binding regions have recently been identified (Parnell et al. 2006; Ramos et al. 2006; Adryan et al. 2007). These non-gypsy regions differ from the gypsy insulator in the number and spacing of the Su(Hw)-binding sites, with single sites dominating. As the generation of an insulator requires a cluster of at least four gypsy insulator Su(Hw) sites (Scott et al. 1999), it is unclear whether all endogenous Su(Hw) regions possess the same properties as the gypsy insulator. Initial studies of the enhancer-blocking capacity of endogenous regions carrying fewer than four Su(Hw) sites demonstrated that these sequences prevent enhancer-activated transcription, but with a reduced strength relative to the gypsy insulator and with a dependence on the genomic integration site, properties not seen for transgenes containing the gypsy insulator (Parnell et al. 2006; Ramos et al. 2006). The exception to these observations was SI 62D, which established a block of the yellow wing and body enhancers as strong as the gypsy insulator (Parnell et al. 2006). For this reason, we continued our studies with SI 62D.
Properties of Su(Hw) association with the gypsy and endogenous insulators differ:
Sequence analysis of SI 62D predicted the presence of four Su(Hw)-binding sites. However, direct tests of Su(Hw) binding in EMSA studies showed that only three were bona fide binding sites (Figure 3). These data imply that, unlike three gypsy insulator sites, three SI 62D Su(Hw)-binding sites confer an enhancer block. We propose that these differences may reflect Su(Hw) protein occupancy at each binding region when these regions are assembled into chromatin. Previous studies have demonstrated that clustering of transcription-factor-binding sites facilitates binding to a chromatin template, as one bound protein appears to weaken interactions between the nucleosome and DNA to reveal the adjacent site (Taylor et al. 1991). We propose that the organization of Su(Hw) sites in the gypsy insulator might act similarly to facilitate DNA binding within a chromatin context. In contrast, endogenous Su(Hw) regions may include binding sites for other proteins that alter local chromatin structure and increase Su(Hw) accessibility to DNA. SI 62D is located within the promoter region of CG32301 (Figure 1), suggesting that promoter factors may enhance in vivo binding of the Su(Hw) protein at this genomic location.
Our studies reveal that in vivo chromatin association of the Su(Hw) protein may be further facilitated by interactions between Su(Hw) and another protein(s). These conclusions are based on our findings that the mutant Su(Hw)f protein bound the gypsy insulator in vitro, even though in vivo binding was lost (Figures 2 and 3). As this mutant protein carries an inactive zinc finger 10, we propose that this finger is required for in vivo association with the gypsy insulator in a sequence-independent manner. It was recently found that E(y2)/Sus1, a component of the SAGA/TFTC histone acetyl transferase complex, binds the Su(Hw) zinc-finger domain (Kurshakova et al. 2007), predicting that this complex may associate with Su(Hw) zinc finger 10 to facilitate Su(Hw) binding to chromosomes. The fact that Su(Hw)f retains in vivo association with SI 62D is consistent with our proposal that these Su(Hw) sites are in a relatively accessible chromatin state.
Enhancer blocking by the gypsy insulator is proposed to result from pairing interactions that form a loop domain, which imposes a physical barrier to communication among enhancers, silencers, and promoters (Kuhn and Geyer 2003; Dorman et al. 2007; Maeda and Karch 2007; Wallace and Felsenfeld 2007). Evidence for insulator interactions comes from studies showing that two gypsy insulators placed between an enhancer and a promoter permit enhancer-activated transcription in a process known as insulator neutralization (Cai and Shen 2001; Muravyova et al. 2001; Kuhn et al. 2003; Majumder and Cai 2003). These gypsy insulator interactions are selective, such that by combining gypsy and a heterologous insulator, such as scs or scs′, enhancer-blocking activity is retained (Kuhn et al. 2003; Majumder and Cai 2003). To gain insights into whether endogenous Su(Hw) sites show insulator interactions, we tested SI 62D in an insulator neutralization assay. We found that neutralization occurred when two SI 62D insulators were paired, but not when SI 62D was paired with the gypsy insulator (Figure 1). These findings are surprising, as both insulators bind the Su(Hw) protein and recruit other gypsy insulator proteins, such as Mod67.2 (Parnell et al. 2006). Two explanations are possible. First, the distinct organization of Su(Hw)-binding sites in SI 62D might inhibit interactions with the gypsy insulator. Alternatively, additional proteins bound to SI 62D might interfere with pairing. Our data are consistent with previous studies showing selective interactions among insulators (Kuhn et al. 2003; Majumder and Cai 2003) and further limit the scope of possible gypsy insulator associations.
The role of the zinc-finger domain in insulator activity:
The zinc-finger domain of the Su(Hw) protein is highly conserved, demonstrating 80% identity and 95% similarity over 40–60 million years of evolution (Harrison et al. 1993). Yet, a role for only 5 of the 12 fingers has been defined (Harrison et al. 1993; Kim et al. 1996). To determine whether these fingers play a role in insulator function other than chromosome association, we tested the ability of zinc-finger domain mutants to block enhancers and a PRE silencer using a Su(Hw)-LexA tethering system. These studies demonstrated that artificially tethered Su(Hw) proteins that either carried a mutated zinc-finger domain or lacked this domain completely were sufficient to prevent both enhancer and silencer modulation of target promoters. These findings imply that the role of Su(Hw) zinc-finger domain is restricted to chromosome association, with some fingers, such as zinc finger 7, required for DNA recognition and another, zinc finger 10, required for sequence-independent in vivo chromosome association. It remains possible that the Su(Hw) protein has multiple cellular activities, with insulator effects representing one type. Future studies are needed to determine whether insulator-independent functions do exist and whether these activities require the highly conserved zinc-finger domain.
Footnotes
These authors contributed equally to this work.
Present address: University of Utah, Salt Lake City, UT 84112.
Footnotes
Communicating editor: K. G. Golic
Acknowledgement
We thank Ivan Clark, Astrid Skjesol, and Daisuke Mayuzumi for technical contributions to this work. We recognize Lori Wallrath and members of the Geyer laboratory for their critical reading of this manuscript. We thank Victor Corces for supplying the Su(Hw) antibody used in the chromatin immunoprecipitation studies. This work was supported by National Institutes of Health Grant GM42539 to P.K.G. and the Carver College of Medicine Collaborative Grant to P.K.G. and M.S.W.
References
Adryan, B., G. Woerfel, I. Birch-Machin, S. Gao, M. Quick et al.,
Brasset, E., and C. Vaury,
Cai, H., and M. Levine,
Cai, H. N., and M. Levine,
Cai, H. N., and P. Shen,
Capelson, M., and V. G. Corces,
Chetverina, D., E. Savitskaya, O. Maksimenko, L. Melnikova, O. Zaytseva et al.,
Chia, W., G. Howes, M. Martin, Y. B. Meng, K. Moses et al.,
Comet, I., E. Savitskaya, B. Schuettengruber, N. Negre, S. Lavrov et al.,
Dorman, E. R., A. M. Bushey and V. G. Corces,
Dorsett, D., G. A. Viglianti, B. J. Rutledge and M. Meselson,
Engel, N., J. L. Thorvaldsen and M. S. Bartolomei,
Festenstein, R., M. Tolaini, P. Corbella, C. Mamalaki, J. Parrington et al.,
Filippova, G. N., S. Fagerlie, E. M. Klenova, C. Myers, Y. Dehner et al.,
Fyrberg, E. A., J. W. Mahaffey, B. J. Bond and N. Davidson,
Gamsjaeger, R., C. K. Liew, F. E. Loughlin, M. Crossley and J. P. Mackay,
Georgiev, P. G., and T. I. Gerasimova,
Gerasimova, T. I., D. A. Gdula, D. V. Gerasimov, O. Simonova and V. G. Corces,
Geyer, P. K., and V. G. Corces,
Geyer, P. K., and V. G. Corces,
Geyer, P. K., M. M. Green and V. G. Corces,
Golovnin, A., I. Birukova, O. Romanova, M. Silicheva, A. Parshikov et al.,
Gruzdeva, N., O. Kyrchanova, A. Parshikov, A. Kullyev and P. Georgiev,
Hagstrom, K., M. Muller and P. Schedl,
Harrison, D. A., M. A. Mortin and V. G. Corces,
Harrison, D. A., D. A. Gdula, R. S. Coyne and V. G. Corces,
Hoover, K. K., T. I. Gerasimova, A. J. Chien and V. G. Corces,
Jakobsson, J., N. Rosenqvist, L. Thompson, P. Barraud and C. Lundberg,
Kim, J., B. Shen, C. Rosen and D. Dorsett,
Kuhn, E. J., and P. K. Geyer,
Kuhn, E. J., M. M. Viering, K. M. Rhodes and P. K. Geyer,
Kurshakova, M., O. Maksimenko, A. Golovnin, M. Pulina, S. Georgieva et al.,
Lobanenkov, V. V., R. H. Nicolas, V. V. Adler, H. Paterson, E. M. Klenova et al.,
Maeda, R. K., and F. Karch,
Majumder, P., and H. N. Cai,
Mallin, D. R., J. S. Myung, J. S. Patton and P. K. Geyer,
Modolell, J., W. Bender and M. Meselson,
Morris, J. R., J. Chen, S. T. Filandrinos, R. C. Dunn, R. Fisk et al.,
Muravyova, E., A. Golovnin, E. Gracheva, A. Parshikov, T. Belenkaya et al.,
Noma, K., C. D. Allis and S. I. Grewal,
Pai, C. Y., E. P. Lei, D. Ghosh and V. G. Corces,
Parkhurst, S. M., D. A. Harrison, M. P. Remington, C. Spana, R. L. Kelley et al.,
Parnell, T. J., M. M. Viering, A. Skjesol, C. Helou, E. J. Kuhn et al.,
Parnell, T. J., E. J. Kuhn, B. L. Gilmore, C. Helou, M. S. Wold et al.,
Peifer, M., and W. Bender,
Pikaart, M. J., F. Recillas-Targa and G. Felsenfeld,
Ramos, E., D. Ghosh, E. Baxter and V. Corces,
Recillas-Targa, F., V. Valadez-Graham and C. M. Farrell,
Roseman, R. R., V. Pirrotta and P. K. Geyer,
Roseman, R. R., E. A. Johnson, C. K. Rodesch, M. Bjerke, R. N. Nagoshi et al.,
Scott, K. C., A. D. Taubman and P. K. Geyer,
Scott, K. S., and P. K. Geyer,
Sigrist, C. J., and V. Pirrotta,
Spana, C., and V. G. Corces,
Spana, C., D. A. Harrison and V. G. Corces,
Taylor, I. C., J. L. Workman, T. J. Schuetz and R. E. Kingston,
Valenzuela, L., and R. T. Kamakaka,
Wallace, J. A., and G. Felsenfeld,
Yannaki, E., J. Tubb, M. Aker, G. Stamatoyannopoulos and D. W. Emery,
Yoon, Y. S., S. Jeong, Q. Rong, K. Y. Park, J. H. Chung et al.,



![ChIP analysis of endogenous Su(Hw) sites in a wild-type and su(Hw) mutant background. (A) PCR analysis. Shown are representative examples of PCR products obtained from chromatin material that was either directly purified (input, or IN) or immunoprecipitated with either Su(Hw) antibody (SH) or a nonspecific rabbit (NS) antibody. Amplified regions correspond to the gypsy insulator, hsp26 coding region, and Su(Hw) insulators 1A-2 and 62D. Chromatin was derived from wild-type [Su(Hw)+/+] and mutant [Su(Hw)v/f] larvae. (B) ChIP analysis of endogenous SIs. The percentage of input was determined by quantifying the intensity of the PCR product in the immunoprecipitated fraction relative to input. Wild-type ratios are indicated by solid bars, while mutant ratios are indicated by shaded bars. The average of at least three ChIP experiments is shown, with error bars indicating the standard deviation.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/genetics/179/3/10.1534_genetics.108.087254/3/m_1263fig2.jpeg?Expires=1716699645&Signature=uG1-N8KrCGTRFRZbGfWd2f3AgBOMtA-U1OYpL~CCyf~hXFIt0H3JZQRTwv5s54vTvoFDPCzb5fjm0C8jpVQQy1PHM7~L2jBwrOX~vEipPHxElj~WNUlSmDwAc-7ldJts9tW-j29wWomVCypivLHUjyj3SylB4qzLuEHedujN4n~IZh-AvuemuBAAU~aUOTFL1TXesFiuAgmw90j7C~0g-ZoYEP6yksAWLt-VEb4xyyseMELTTZOEQAetoXQVgXJc48KEUhOBWoV6qFVAEpLH4VBsPSyUjlsAkHlKqShRTK~vOLwcu1Ds3lAZin5gygtEcghZ8ge7ld6ALLZMXDc5Eg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Summary of results obtained in the LexA-tethering assay. (Left) Percentage of lines that reconstituted enhancer blocking (top) and silencing blocking (bottom) using the yellow gene as a reporter. The numbers in parentheses represent the lines showing insulator activity relative to the total number tested. (Middle) Structures of the two reporter transposons. P[y L En L w] contains a LexA BR (L in shaded box) of six repeated sites inserted between the yellow wing and body enhancers (ovals marked W and B) and yellow (y) promoter and between the eye enhancer (oval marked E) and white promoter (w). A gypsy insulator (inverted solid triangle) is located at the 3′-end of the white gene to prevent position effects. P[y PRE L w] contains the LexA BR (L in shaded boxes) inserted between the white eye enhancer and the PRE. Solid boxes indicate P-element ends. (Right) Percentage of lines that reconstituted enhancer blocking (top) and silencing blocking (bottom) using the white gene as a reporter.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/genetics/179/3/10.1534_genetics.108.087254/3/m_1263fig5.jpeg?Expires=1716699645&Signature=VxJtHB402p7rrk8-aXJLQFAWNwgcxufFFnTc0mMfxcbloRNeH1us2w8huXQS82mHfeNjEyk87v1vXFyd9XoZMt0ZJ0xKMWBnRscBPDFxX~KsIEo1OOIlLEd60hPuJX6tSQEyvy5gV2nCcuU-ZeUc59tDZzE3xXTSe1pKUJbWFy8EqmzZe4~aZVWKU4uUQX9h5L1W9po7ToMRXUL4BP8yPBWpPxmQfNy3ALBpfQP5oFDuPAyrvz9M~akcKoaAUVT-9JGJDQiNoi3Ra7U9OSJpqa442c8bOzjG6dP-hafOGjF1dw3jD0NMxCa-sTd87pp~jJMuHQIRM70prsWW4rejvw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
