-
PDF
- Split View
-
Views
-
Cite
Cite
Soochin Cho, Zachary Y Huang, Jianzhi Zhang, Sex-Specific Splicing of the Honeybee doublesex Gene Reveals 300 Million Years of Evolution at the Bottom of the Insect Sex-Determination Pathway, Genetics, Volume 177, Issue 3, 1 November 2007, Pages 1733–1741, https://doi.org/10.1534/genetics.107.078980
Close - Share Icon Share
Abstract
Sex-determination mechanisms vary greatly among taxa. It has been proposed that genetic sex-determination pathways evolve in reverse order from the final step in the pathway to the first step. Consistent with this hypothesis, doublesex (dsx), the most downstream gene in the Drosophila sex-determination cascade that determines most sexual phenotypes also determines sex in other dipterans and the silk moth, while the upstream genes vary among these species. However, it is unknown when dsx was recruited to the sex-determination pathway during insect evolution. Furthermore, sex-specific splicing of dsx, by which dsx determines sex, is different in pattern and mechanism between the moth and the fly, raising an interesting question of how these insects have kept the executor of sex determination while allowing flexibility in the means of execution. To address these questions, here we study the dsx gene of the honeybee Apis mellifera, a member of the most basal lineage of holometabolous insects. We report that honeybee dsx is sex-specifically spliced and that it produces both the fly-type and moth-type splicing forms, indicating that the use of different splicing forms of Dsx in controlling sexual differentiation was present in the common ancestor of holometabolous insects. Our data suggest that in ancestral holometabolous insects the female Dsx form is the default and the male form is generated by suppressing the splicing of the female form. Thus, it is likely that the dsx splicing activator system in flies, where the male form is the default, arose during early dipteran evolution.
THE genetic basis underlying sex-determination evolution is a fascinating topic because sex-determination mechanisms vary greatly across different species (Bull 1983; Marin and Baker 1998). It was postulated a decade ago that genetic sex-determination pathways evolve in reverse order from the final step in the hierarchy up to the first (Wilkins 1995). This hypothesis has reaped supporting evidence from studies of multiple model organisms, including invertebrate and vertebrate animals. One of the most fruitful areas is the sex-determination pathway of the fruit fly Drosophila melanogaster and its related dipteran species (Grahamet al. 2003; Pomiankowskiet al. 2004). In Drosophila, the ratio between the number of X chromosomes and autosomal sets (X:A) serves as the initial determinant of sex. In females, the X:A ratio of 1 activates the Sex lethal (Sxl) gene that encodes a splicing regulator. Sxl protein then activates the female-specific splicing of its downstream gene transformer (tra), giving rise to functional Tra protein. Tra, together with Tra2, activates the female-specific splicing of its downstream gene doublesex (dsx), generating the female-type Dsx protein (DsxF), which regulates its downstream genes for the female development to proceed. In males, the X:A ratio of 0.5 prevents Sxl from being produced, leading to the male-specific splicing of tra, rendering its encoded product nonfunctional due to a premature stop codon. In the absence of Tra activity, DsxM is produced by the default male-type splicing of dsx, which regulates its downstream genes for male development (Cline and Meyer 1996).
Interestingly, in non-Drosophila insects, Sxl is expressed in both sexes equally, suggesting that the sex-determining role of Sxl is limited to Drosophila (Meiseet al. 1998; Sacconeet al. 1998; Sievertet al. 2000; Lagoset al. 2005; Niimiet al. 2006). In line with this observation, it was found that Sxl was duplicated in the brachyceran dipteran (fly) lineage after its separation from nematoceran dipterans (mosquitoes, gnats, and midgets), which may have contributed to the adoption of Sxl to the sex-determination pathway in Drosophila (Trautet al. 2006). In the olive fruit fly (Bactrocera oleae) and Mediterranean fruit fly (Ceratitis capitata), two non-Drosophila flies, tra was found to activate the female-specific splicing of dsx as in Drosophila, but the TRA activity in the female is maintained by an autoregulatory loop, unlike in Drosophila where it is regulated by Sxl (Paneet al. 2002, 2005; Lagoset al. 2007).
Dsx, the most downstream component of the Drosophila sex-determination pathway that controls most sex-specific phenotypes, is a transcriptional factor with a zinc-finger DNA-binding domain known as the DM domain. Several DM-domain-containing proteins are known to participate in sex determination in a diverse array of animals. These genes include mab-3, the male somatic sex-determining gene in nematodes (Raymondet al. 1998); DMY, the master regulator of male development in medaka fish (Matsudaet al. 2002; Nandaet al. 2002; Zhang 2004); and Dmrt1, the gene required for mammalian testis differentiation (Raymondet al. 2000). However, unlike dsx, these genes do not use sex-specific splicing to determine sex, and their sequence similarity with dsx is limited to the DM domain, indicating that they may not be orthologous to dsx. Orthologs of D. melanogaster dsx have been identified and studied in a number of dipterans (Shearman and Frommer 1998; Kuhnet al. 2000; Hedigeret al. 2004; Lagoset al. 2005; Ruizet al. 2005; Scaliet al. 2005). In all these species, the gene structure and the sex-specific splicing pattern of dsx are generally conserved. Furthermore, as in D. melanogaster (Inoueet al. 1992), it appears that the Tra/Tra2 complex binds to the cis-regulatory element (dsxRE) within the female-specific exon, activating the weak 3′ splicing site preceding the exon to give rise to the female-type dsx mRNA. Outside the order Diptera, relatively detailed studies of dsx have been conducted in the silk moth Bombyx mori (Ohbayashiet al. 2001; Suzukiet al. 2001, 2003, 2005; Funagumaet al. 2005). Interestingly, the female splicing pattern of dsx in Bombyx is different from that in Drosophila at the 3′-end of the mRNA. In Bombyx, the 3′ splicing site preceding the upstream female-specific exon does not appear to be weak and dsxRE has not been found within the female-specific exons. In addition, in vitro splicing experiments showed that, unlike in Drosophila, the female-type splicing in Bombyx is the default, suggesting that a different molecular mechanism involving splicing repressors, rather than activators, is used to generate sex-specific variants of silk moth dsx mRNA (Suzukiet al. 2001), although this interpretation needs further scrutiny because the in vitro splicing experiment for the moth dsx was conducted in HeLa cell extract, which may be different from the insect cellular environment. The disparity between the fly and moth systems raises an intriguing evolutionary question of which system is ancestral and which is derived. It is also unknown whether the use of dsx in sex determination is conserved outside the dipterans and lepidopterans.
The honeybee Apis mellifera is an ideal organism for addressing the above questions because of its phylogenetic position within holometabolous insects, which are insects that undergo a complete cycle of metamorphism. Recent studies aided by the completion of the honeybee genome sequencing revealed that the order Hymenoptera, which includes the honeybee, is the most basal lineage in the phylogeny of holometabolous insects (superorder Endopterygota), being an outgroup to Diptera, Lepidoptera, and Coleoptera (HoneybeeGenomeSequencingConsortium 2006; Savardet al. 2006). Understanding the sex-determination mechanism in honeybees will thus shed light on the evolution of dsx in holometabolous insects and help to further verify Wilkins's hypothesis on the evolution of sex-determination pathways. Honeybees, as well as >100,000 species in the insect order of Hymenoptera, are haplodiploids (Cook 1993). Fertilized eggs of honeybees become diploid females (workers and queens) and unfertilized eggs parthenogenically develop as haploid males (drones). More specifically, honeybee sex is controlled by the allelic composition of a locus known as csd (complementary sex determination). Diploid individuals carrying two different csd alleles (heterozygotes) develop as females, while haploid individuals become males. Diploid embryos homozygous for csd become sterile males. As expected, csd is subject to balancing selection and is highly polymorphic (Hasselmann and Beye 2004; Choet al. 2006). The csd gene encodes a member of the SR protein family, whose members are involved in splicing regulation of various mRNAs; however, the genes that are regulated by Csd are unknown (Beyeet al. 2003). On the basis of protein sequence comparison, Csd appears to be homologous to D. melanogaster Tra protein (Beyeet al. 2003), which plays a role in sex determination by regulating sex-specific splicing of its downstream gene dsx. Therefore, one of the potential targets of Csd is dsx. Very recently, Cristinoet al. 2006 identified a dsx homolog in the honeybee genome sequence using computer-based prediction and demonstrated that a pair of primers designed from their gene prediction could amplify a part of dsx cDNA only from males, but not from females, suggesting that the honeybee dsx is sex-specifically spliced. However, they did not obtain full-length dsx mRNA sequences from the two sexes and thus the crucial information on the sex-specific splicing pattern and mechanism is missing. In this study, we identify four different splicing variants of dsx mRNA in honeybees: two female specific, one male specific, and one ubiquitous. We also determine the full-length mRNA sequences and the complete gene structure. Our comparative analysis suggests that (1) sexual differentiation regulated by sex-specific splicing of dsx was present in the common ancestor of holometabolous insects and that (2) the default dsx splicing form switched from female in ancestral holometabolous insects to male in dipterans, with the alternative splicing mechanism changing from repressing the default to activating the alternative form.
MATERIALS AND METHODS
Bee sampling, RNA purification, and dsx mRNA sequence determination:
Honeybee A. mellifera (Am for short) worker and drone eggs, young larvae (1–3 days after hatching), old larvae (prepupae), and pupae were sampled directly from colonies located at East Lansing, Michigan. The samples were immediately frozen in a dry-ice chamber and stored in a −70° freezer until used. Total RNA was purified from each bee sample using TRIZOL reagent (Invitrogen, Carlsbad, CA). To determine the entire sequences of the Am-dsx transcripts contained in the total RNA samples, 5′ and 3′ rapid amplification of cDNA ends (RACE) was performed using the FirstChoice RLM-RACE kit (Ambion, Austin, TX). Gene-specific primers for RACE reactions were designed referring to a partial cDNA sequence available from GenBank (accession no. AY375535) (Table 1). All RACE products were cloned into pCR2.1 vector (Invitrogen) before being sequenced by the conventional dideoxy method at the University of Michigan DNA Sequencing Core.
Primers used in this study
Name . | Sequence (from 5′ to 3′) . | Target exon . |
|---|---|---|
| For RACE | ||
| AmdsxF1 | CGCACAAGAGGTACTGCAAG | 2 |
| AmdsxF2 | CCTGCGAGAAGTGTAAGATCAC | 2 |
| AmdsxR1 | GACTTCTAGGTGGTTGAGGGAC | 3 |
| AmdsxR2 | CTTTATCCTGTGCCAGGTGTC | 2 |
| For RT–PCR | ||
| AmdsxF3 | CATGTCGGCTTGTCTACGCAC | 1 |
| AmdsxR3 | CGATGTAATCATTCCTCTTTGG | 7 |
| AmdsxR4 | CAAACAACCATTCGTGAAAATCTC | 5 |
| AmdsxR5 | CGCCAAGAAACCATTATGAAAC | 2 |
| AmgapdhFa | GTTCGGTGCTCAGGTTGTTG | — |
| AmgapdhRa | CCAGCTTGTAAATGACCAGAAGC | — |
Name . | Sequence (from 5′ to 3′) . | Target exon . |
|---|---|---|
| For RACE | ||
| AmdsxF1 | CGCACAAGAGGTACTGCAAG | 2 |
| AmdsxF2 | CCTGCGAGAAGTGTAAGATCAC | 2 |
| AmdsxR1 | GACTTCTAGGTGGTTGAGGGAC | 3 |
| AmdsxR2 | CTTTATCCTGTGCCAGGTGTC | 2 |
| For RT–PCR | ||
| AmdsxF3 | CATGTCGGCTTGTCTACGCAC | 1 |
| AmdsxR3 | CGATGTAATCATTCCTCTTTGG | 7 |
| AmdsxR4 | CAAACAACCATTCGTGAAAATCTC | 5 |
| AmdsxR5 | CGCCAAGAAACCATTATGAAAC | 2 |
| AmgapdhFa | GTTCGGTGCTCAGGTTGTTG | — |
| AmgapdhRa | CCAGCTTGTAAATGACCAGAAGC | — |
These primers amplify the glyceraldehyde 3 phosphate dehydrogenase 1 gene used as the positive control.
Primers used in this study
Name . | Sequence (from 5′ to 3′) . | Target exon . |
|---|---|---|
| For RACE | ||
| AmdsxF1 | CGCACAAGAGGTACTGCAAG | 2 |
| AmdsxF2 | CCTGCGAGAAGTGTAAGATCAC | 2 |
| AmdsxR1 | GACTTCTAGGTGGTTGAGGGAC | 3 |
| AmdsxR2 | CTTTATCCTGTGCCAGGTGTC | 2 |
| For RT–PCR | ||
| AmdsxF3 | CATGTCGGCTTGTCTACGCAC | 1 |
| AmdsxR3 | CGATGTAATCATTCCTCTTTGG | 7 |
| AmdsxR4 | CAAACAACCATTCGTGAAAATCTC | 5 |
| AmdsxR5 | CGCCAAGAAACCATTATGAAAC | 2 |
| AmgapdhFa | GTTCGGTGCTCAGGTTGTTG | — |
| AmgapdhRa | CCAGCTTGTAAATGACCAGAAGC | — |
Name . | Sequence (from 5′ to 3′) . | Target exon . |
|---|---|---|
| For RACE | ||
| AmdsxF1 | CGCACAAGAGGTACTGCAAG | 2 |
| AmdsxF2 | CCTGCGAGAAGTGTAAGATCAC | 2 |
| AmdsxR1 | GACTTCTAGGTGGTTGAGGGAC | 3 |
| AmdsxR2 | CTTTATCCTGTGCCAGGTGTC | 2 |
| For RT–PCR | ||
| AmdsxF3 | CATGTCGGCTTGTCTACGCAC | 1 |
| AmdsxR3 | CGATGTAATCATTCCTCTTTGG | 7 |
| AmdsxR4 | CAAACAACCATTCGTGAAAATCTC | 5 |
| AmdsxR5 | CGCCAAGAAACCATTATGAAAC | 2 |
| AmgapdhFa | GTTCGGTGCTCAGGTTGTTG | — |
| AmgapdhRa | CCAGCTTGTAAATGACCAGAAGC | — |
These primers amplify the glyceraldehyde 3 phosphate dehydrogenase 1 gene used as the positive control.
Reverse transcription polymerase chain reaction and gel electrophoresis:
Total RNA samples were reverse transcribed using the RETROscript kit (Ambion), and PCR was performed for each cDNA sample using Platinum Taq DNA polymerase (Invitrogen) with the gene-specific primers listed in Table 1. The PCR cycles used were 95° for 2 min, then 35 cycles of 30 sec at 95°, 30 sec at 58°, and 2 min at 72°, followed by final synthesis at 72° for 7 min. PCR amplicons were analyzed by 1% agarose gel electrophoresis and verified by DNA sequencing.
Sequence analysis:
The exon–intron boundaries of honeybee dsx were determined by comparing the cDNA sequences from this study and the genomic DNA sequence (accession no. NW_001253366) generated by the honeybee genome project (http://www.ncbi.nlm.nih.gov/genome/guide/bee/). The mRNA sequences of the dsx genes in D. melanogaster (Dm-dsx), the mosquito Anopheles gambiae (Ag-dsx), and the silk moth B. mori (Bm-dsx) were obtained from GenBank and their accession numbers are listed in Table 2. We used the UCSC Genome Bioinformatics site (http://genome.ucsc.edu/) to obtain the genomic sequences corresponding to these mRNA sequences to determine their gene structures. For A. gambiae dsx, two independent groups submitted their sequences to GenBank: AY903307 (Ag-dsxM) and AY903308 (Ag-dsxF) and DQ137801 (Ag-dsxM) and DQ137802 (Ag-dsxF). We used the former pair in this study because (1) we found that the latter pair had a large number of mismatches in the coding region compared with the completed genome sequence and (2) the 5′ and 3′ untranslated regions (UTRs) in the latter pair were not alignable with the genome sequence. Protein and nucleotide sequences were aligned by Clustal X (Thompsonet al. 1997) with manual adjustments.
Holometabolous insect dsx gene sequences used in this study
Species . | Order . | Sex . | GenBank accession no. . |
|---|---|---|---|
| D. melanogaster (fruit fly) | Diptera | Female | NM_169203 |
| Male | NM_169202 | ||
| A. gambiae (mosquito) | Diptera | Female | AY903308 |
| Male | AY903307 | ||
| B. mori (silk moth) | Lepidoptera | Female | AB048543 |
| Male | AB048544 | ||
| A. mellifera (honeybee) | Hymenoptera | Female | EU137059, EU137060 (this study) |
| Male | EU137057 (this study) |
Species . | Order . | Sex . | GenBank accession no. . |
|---|---|---|---|
| D. melanogaster (fruit fly) | Diptera | Female | NM_169203 |
| Male | NM_169202 | ||
| A. gambiae (mosquito) | Diptera | Female | AY903308 |
| Male | AY903307 | ||
| B. mori (silk moth) | Lepidoptera | Female | AB048543 |
| Male | AB048544 | ||
| A. mellifera (honeybee) | Hymenoptera | Female | EU137059, EU137060 (this study) |
| Male | EU137057 (this study) |
Holometabolous insect dsx gene sequences used in this study
Species . | Order . | Sex . | GenBank accession no. . |
|---|---|---|---|
| D. melanogaster (fruit fly) | Diptera | Female | NM_169203 |
| Male | NM_169202 | ||
| A. gambiae (mosquito) | Diptera | Female | AY903308 |
| Male | AY903307 | ||
| B. mori (silk moth) | Lepidoptera | Female | AB048543 |
| Male | AB048544 | ||
| A. mellifera (honeybee) | Hymenoptera | Female | EU137059, EU137060 (this study) |
| Male | EU137057 (this study) |
Species . | Order . | Sex . | GenBank accession no. . |
|---|---|---|---|
| D. melanogaster (fruit fly) | Diptera | Female | NM_169203 |
| Male | NM_169202 | ||
| A. gambiae (mosquito) | Diptera | Female | AY903308 |
| Male | AY903307 | ||
| B. mori (silk moth) | Lepidoptera | Female | AB048543 |
| Male | AB048544 | ||
| A. mellifera (honeybee) | Hymenoptera | Female | EU137059, EU137060 (this study) |
| Male | EU137057 (this study) |
RESULTS
Molecular cloning of full-length Am-dsx cDNAs from males and females:
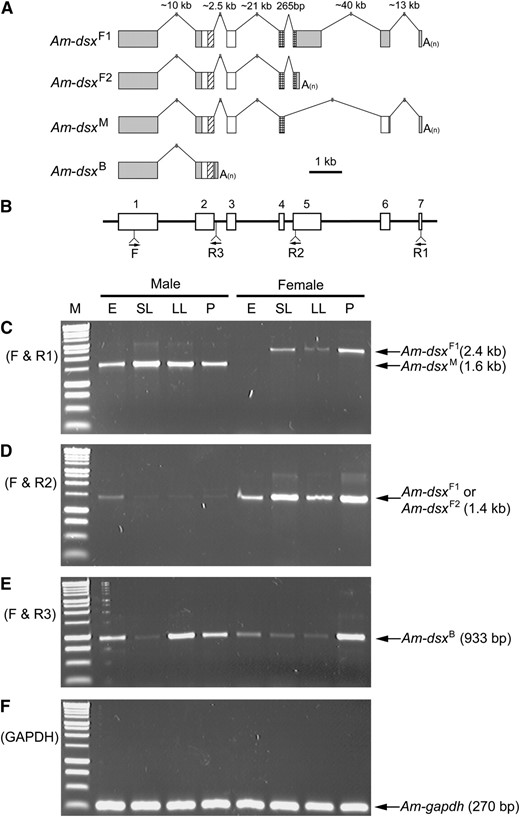
To determine the full-length coding and 5′- and 3′-UTR sequences of the honeybee dsx gene, we performed RACE using total RNA samples purified from four different developmental stages of female workers and male drones: eggs, young larvae, old larvae, and pupae. Gene-specific primers for both 5′ and 3′ RACE experiments were designed to target the exon that harbors the conserved DNA-binding domain and the predicted start codon. From our 5′ RACE in both sexes, we identified another exon of ∼1.2 kb upstream from this predicted exon. Because no open reading frame of significant length was identified within the new exon, we regarded it as noncoding. From our 3′ RACE, we detected four variant transcripts: two female specific, one male specific, and one from both sexes (Figure 1A). The longer female-specific variant (dsxF1, 3359 nucleotides) contains seven exons, and the shorter (dsxF2, 2337 nucleotides) contains five. The difference between dsxF1 and dsxF2 rests entirely in the 3′-UTR, owing to alternative transcription termination sites. As a result, dsxF1 and dsxF2 encode the same protein (DsxF) of 277 amino acids. The sole male-specific transcript (dsxM, 2504 nucleotides) has six exons, skipping the fifth exon of dsxF by alternative splicing. Consequently, dsxM encodes a protein product (DsxM) of 336 amino acids that is identical to DsxF at its N-terminal 250 amino acids but different at the C-terminal part (Figure 1A; supplemental Figure 1 at http://www.genetics.org/supplemental/). Finally, a fourth variant (dsxB, 1992 nucleotides) detected from both sexes shares its first exon with the other variants, but its second exon is extended by 217 nucleotides to the 3′-end compared with the other variants, overlooking the 5′ splicing site at the end of the second exon (Figure 1A). This shortest variant encodes a protein of 130 amino acids (DsxB), whose last six amino acids are different from DsxF and DsxM (Figure 1A; supplemental Figure 1 at http://www.genetics.org/supplemental/).
Gene structure and expression pattern of the four splicing variants of honeybee dsx. (A) Structures of the four splicing variants. Boxes represent exons and lines are introns. Only exons are drawn to scale. Shaded boxes are untranslated regions and open boxes are coding regions. Within coding regions, diagonally hatched boxes mark oligomerization domain 1/DNA-binding domain and cross-checked boxes mark oligomerization domain 2. “A(n)” represents the polyadenylated tail. (B) The locations of the primers used for RT–PCR experiments are shown as arrows. Open boxes are exons and lines are introns. They are drawn in alignment with the gene structures in A. The only forward primer (F) was designed to anneal to the first exon, which is shared by all four variants. R1 anneals to dsxF1 and dsxM, R2 to dsxF1 and dsxF2, and R3 to only dsxB. (C–F) RT–PCR amplicons from total RNA samples of four different developmental stages of both sexes are analyzed by 1% agarose gel electrophoresis. E, embryos; SL, small larvae; LL, large larvae; P, pupae. See materialsandmethods for detailed information on the samples. The leftmost lane (M) is the size standard (BenchTop 1-kb DNA ladder, Promega, Madison, WI). Names and sizes of amplicons are labeled with arrows. Primers F and R1 were used for C, F and R2 for D, and F and R3 for E. Primers for the honeybee glyceraldehyde 3-phosphate dehydrogenase 1 gene were used as positive controls for F.
Sex-specific expression patterns of Am-dsx mRNA variants:
We performed reverse transcription polymerase chain reaction (RT–PCR) to determine the expression patterns of these four variants using one specific forward primer (F) and three reverse primers (R1, R2, and R3) (Figure 1B). Primers F and R1 amplified dsxF1 and dsxM from female and male samples, respectively, in a sex-specific manner (Figure 1C). While dsxM is expressed throughout the male developmental stages tested here, dsxF1 is not expressed in female eggs. Primers F and R2 amplified dsxF1 and dsxF2 from female samples at all stages, but these primers also detected relatively weak expressions of these female variants in males, particularly during the egg stage, suggesting that the female variants are present in male tissues at a low level (Figure 1D). By contrast, the male-specific variant dsxM was not detected in female samples even at a low level (Figure 1C). We confirmed that primers F and R3 specifically amplified dsxB in both sexes (Figure 1E). As a positive control, we used the glyceraldehyde 3-phosphate dehydrogenase 1 gene (GenBank accession no. XM_623046), which showed a constitutive expression throughout all developmental stages in both sexes (Figure 1F).
The C-terminal part of the second oligomerization domain is not conserved in Am-Dsx:
In D. melanogaster, the Dsx protein has two domains (OD1 and OD2) known to be important for oligomerization (Anet al. 1996). OD1 also encompasses the zinc-finger DNA-binding domain (DBD), which is necessary for binding to its target sequences for transcriptional regulation (Erdman and Burtis 1993). The N-terminal part of OD2 is shared by Dm-DsxM and Dm-DsxF, but the C-terminal 15 amino acids are encoded by a female-specific exon and are absent in Dm-DsxM (Anet al. 1996). Previous studies demonstrated that OD1/DBD and OD2 domains are evolutionarily well conserved among all Dsx homologs found in dipterans and the silk moth (Shearman and Frommer 1998; Kuhnet al. 2000; Suzukiet al. 2001; Hedigeret al. 2004; Lagoset al. 2005; Ruizet al. 2005; Scaliet al. 2005). In Am-Dsx, OD1/DBD is conserved with all of the six amino acid residues important for its zinc-finger function unchanged (Erdman and Burtis 1993) (Figure 2A). The N-terminal part of OD2 shared by Am-DsxF and Am-DsxM is also relatively well conserved in evolution (Figure 2A). However, the C-terminal part of OD2 specific to DsxF is not conserved in Am-DsxF, with only 1 of the 15 amino acids in the region identical to Dm-DsxF (Figure 2B). It is known that the level of sequence identity in the male-specific part of DsxM declines rapidly when Dm-DsxM is compared with other species beyond the genus level (Shearman and Frommer 1998; Kuhnet al. 2000; Hedigeret al. 2004; Lagoset al. 2005). As expected, this region of Am-DsxM does not show any significant sequence similarity to the DsxM proteins in other insects (Figure 2C).
Amino acid sequence alignment of Dsx proteins of D. melanogaster (Dm), A. gambiae (Ag), B. mori (Bm), and A. mellifera (Am). Dashes show alignment gaps and dots represent the same amino acids as in Dm-Dsx. Oligomerization domain 1/DNA-binding domain (OD1/DBD) and oligomerization domain 2 (OD2) are boxed. The six amino acids important for binding to the DNA target are labeled with an asterisk. The locations of introns are indicated by numbers; numbers in parentheses indicate the phase of the intron. The numbering of amino acids indicated at the right is according to that of Dm-Dsx. The part shared by both male and female types is shown in A, the female-specific part is shown in B, and the male-specific parts are shown in C.
Variation of dsx splicing patterns in holometabolous insects:
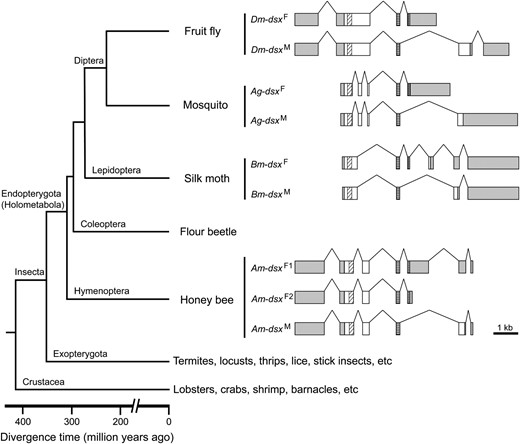
We compared the sex-specific splicing patterns of dsx among D. melanogaster, the mosquito A. gambiae, the moth B. mori, and A. mellifera in the context of the insect phylogeny (Figure 3). In D. melanogaster, the male-specific dsx mRNA connects exons 1, 2, 3, 5, and 6, while the female-specific exon 4 is adjoined after exons 1, 2, and 3 in females (Burtis and Baker 1989). Thus, the male and female variants do not share the final exon. The same splicing pattern is observed in A. gambiae (Figure 3). However, B. mori dsx has a different splicing pattern: two female-specific exons are skipped in the male variant (Bm-dsxM) such that the final two exons are shared by both sexes (Suzukiet al. 2001). In A. mellifera, the single female-specific exon in Am-dsxF1 is skipped in the male variant (Am-dsxM), resulting in the final two exons being shared between Am-dsxF1 and Am-dsxM, a situation similar to that observed in B. mori. However, Am-dsxF2 and Am-dsxM differ by alternatively splicing to sex-specific exons at the 3′-end and they thus do not share the 3′ exon, reminiscent of the two dipteran species, D. melanogaster and A. gambiae (Figure 3). On the basis of the parsimony principle, one may infer from the above observations that (1) the common ancestor of holometabolous insects had both the dipteran-type and lepidopteran-type sex-specific dsx splicing patterns, (2) hymenopterans retain both ancestral patterns, and (3) dipterans and lepidopterans each lost one of the ancestral patterns. However, it should be noted that no significant sequence conservation is detected around the 3′-end of the female-specific exon among these insect species. This is not unexpected because in all these species the 3′-end of the female-specific exon is located in untranslated regions, which are usually evolutionarily unconserved.
Gene structure alignment of the dsx genes in four holometabolous insect species. Boxes represent exons and lines represent introns. Only exons are drawn in scale. Shaded boxes are untranslated regions and open boxes are coding regions. Within coding regions, diagonally hatched boxes mark OD1/DBD and cross-checked boxes mark OD2. Note that the exons for the shared and female-specific coding regions are aligned. Untranslated regions and male-specific coding regions could not be aligned due to sequence divergence. The phylogenetic relationships among the four species and major outgroup lineages are shown at the left of the gene structures.
DISCUSSION
In this study, we identified four alternatively spliced full-length cDNA sequences of the honeybee dsx gene. The presence of four dsx splicing variants is unexpected because only two or three have been identified in other species. Among the four honeybee dsx splicing variants, one (dsxB) is expressed in both sexes, one (dsxM) is expressed in males, and two (dsxF1 and dsxF2) are expressed in females. Because the two female splicing variants are identical to each other in protein sequence, each sex produces one sex-specific Dsx protein as in other species. The function of DsxB is enigmatic, because it has only one, instead of two, oligomerization domains, in addition to the DNA-binding domain (supplemental Figure 1 at http://www.genetics.org/supplemental/). Furthermore, DsxB is unlikely to be involved in sex determination because of its expression in both sexes.
What is the molecular mechanism of the sex-specific splicing of honeybee dsx? In D. melanogaster, the default splicing is the male form, connecting exons 1, 2, 3, 5, and 6. The 3′ splice site preceding the female-specific exon (exon 4) contains a sequence of purine nucleotides, which make it a weak splicing acceptor that is overlooked by the spliceosomal machinery in males. In females, the Tra/Tra2 complex binds to several 13-nucleotide splicing enhancer elements (dsxREs) and the purine-rich elements (PREs) present within the female-specific exon to activate the 3′ splice site and produce female-specific mRNA connecting exons 1, 2, 3, and 4 (Inoueet al. 1992; Lynch and Maniatis 1995). In A. gambiae, the same sex-specific splicing pattern is observed (Figure 3), and dsxRE and PRE are also detected in the female-specific exon, suggesting that a similar molecular mechanism is used to generate sex-specific variants in this species (supplemental Figure 2 at http://www.genetics.org/supplemental/). However, Suzukiet al. (2001) showed that B. mori dsx has a sex-specific splicing pattern different from that of D. melanogaster (Figure 3). Although the difference in splicing pattern among these species rests on whether the 3′-end of the female-specific exon is defined by a 5′ splicing site to connect to its following exon or by a cleavage/polyadenylation signal and does not necessarily imply a difference in regulatory mechanism, the subsequent work using in vitro splicing experiments with HeLa cell extract showed that the female type is generated as the default in B. mori (Suzukiet al. 2001). Furthermore, no dsxRE or PRE are found in the female-specific exons, and the 3′ splice site of the intron preceding the female-specific exons does not appear to be weak (Suzukiet al. 2001). Together, Suzukiet al.'s findings suggest that splicing repressors, rather than activators, are turned on in males to suppress the female-specific splicing to generate the male-specific splicing variant. Unlike other insects, honeybees have two female-specific splicing variants that differ in the choice of transcription termination sites (Figure 1A and Figure 3). Interestingly, one of the seven exons of Am-dsxF1 is skipped in the male-specific variant (Am-dsxM), which is reminiscent of Bm-dsx. Nonetheless, the other female-specific variant (Am-dsxF2) is different from Am-dsxM at the 3′-end, similar to the situation in D. melanogaster and A. gambiae. Is the molecular mechanism underlying the honeybee sex-specific splicing of dsx more similar to that of D. melanogaster or to that of B. mori? We believe that it is more likely to be similar to that of B. mori, although the other possibility cannot be excluded completely, as discussed below.
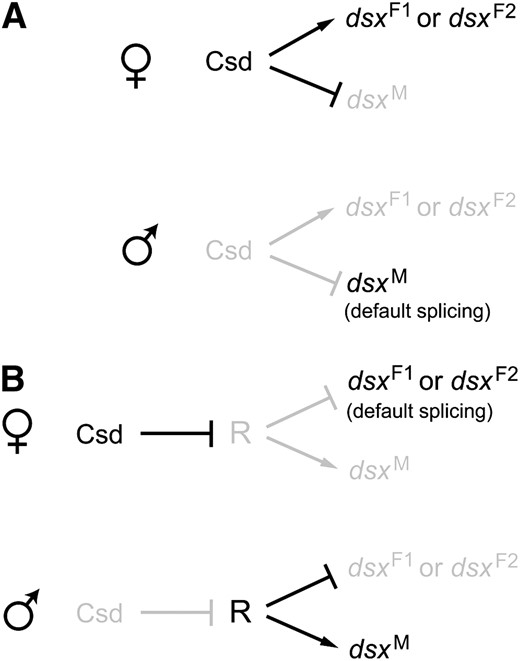
The allelic composition of the csd gene is the master controller of sex differentiation in honeybees; csd heterozygotes develop into females, while homozygotes and hemizygotes develop into males. If honeybees have a fly-type dsx splicing mechanism, the male-specific splicing should be the default and the functional Csd in females directly or indirectly activates the 3′ splice site of the female-specific exon to generate Am-dsxF1 or Am-dsxF2 (Figure 4A). Alternatively, honeybees could have a moth-type splicing mechanism. That is, the female-specific splicing is the default and Csd suppresses splicing repressors in females. In males, the lack of functional Csd leads to the activation of the splicing repressors, which causes the skip of the female-specific exons in splicing and the production of Am-dsxM (Figure 4B). We favor the second scenario for three reasons. First, as is the case of Bm-dsx, no dsxRE or PRE is found in the female-specific exon of Am-dsxF1 or Am-dsxF2. Second, the 3′ splice site preceding the female-specific exon (5′-…ctttattctctag-3′) has only a few purine nucleotides and thus does not appear to be weakened. Third, our RT–PCR experiments detected a low level of female-type dsx expression in male samples but no male type in any female samples, suggesting that the female-type splicing is more likely to be the default (Figure 1, C and D). However, it is still possible that honeybees employ a splicing activator system that is very different from that of D. melanogaster. Experimental investigation, such as in vitro splicing, is required to resolve the two scenarios. If the second scenario involving the repressor system is true, it would be most parsimonious to hypothesize that the common ancestor of all holometabolous insects used a repressor system and the switch to an activator system occurred during dipteran evolution in the common ancestor of fruit flies and mosquitoes. Studies on dsx in other holometabolous insects, such as Tribolium castaneum (order Coleoptera), could further test this idea.
Two models of the genetic regulation of honeybee dsx splicing. Arrows represent activation and lines with a bar indicate suppression. Active genes and interactions are in black whereas inactive ones are in gray. (A) The activator model, where the male form of dsx splicing is the default. In females, functional Csd either directly or indirectly activates the female-specific splicing to give rise to Am-dsxF1 or Am-dsxF2. (B) The repressor model. In females, functional Csd suppresses or inactivates R to allow the default female-type splicing to take place, whereas in males the presumed repressor R suppresses the splicing of the female-specific exon to generate the male-type mRNA, Am-dsxM.
Despite the differences in certain details of the pattern and mechanism, the general similarity in sex-specific splicing of dsx between the honeybee and those insects (Drosophila and Bombyx) where the role of dsx in sex determination has been well established strongly supports the hypothesis that dsx is involved in honeybee sex determination. The final proof of this hypothesis requires showing relevant phenotypes in honeybees where dsx is nonfunctional, which may be possible by RNA interference experiments. If the above hypothesis is correct, as is very likely, our results suggest that the use of alternative splicing of dsx in regulating insect sex differentiation was already present in the common ancestor of holometabolous insects, which existed ∼300 million years ago (Savardet al. 2006). In contrast to dsx, which is at the bottom of the sex-determination pathway, a diverse array of upstream genes and signals in the sex-determination cascade is used in D. melanogaster (X:A ratio), B. mori (a dominant feminizing factor on W), and A. mellifera (complementary allelic composition of a gene). These findings are in strong support of the postulation that genetic sex-determination pathways evolve in reverse order from the final step in the hierarchy up to the first (Wilkins 1995). In the future, it would be interesting to study how sex-specific splicing of honeybee dsx is regulated by upstream genes, such as csd; how differentiation of honeybee sexual traits, in both morphology and behavior, are controlled by the sex-specific Dsx proteins; and whether dsx is also part of the sex-determination pathway in nonholometabolous insects.
Footnotes
Footnotes
Communicating editor: J. A. Lopez
Acknowledgement
We thank Wendy Grus, Wenfeng Qian, and Xiaoxia Wang for valuable comments. This work was supported by a grant from the Office of Vice President for Research at the University of Michigan and by grants from the National Institutes of Health to J.Z.
References
An, W., S. Cho, H. Ishii and P. C. Wensink,
Beye, M., M. Hasselmann, M. Fondrk, R. Page and S. Omholt,
Burtis, K. C., and B. S. Baker,
Cho, S., Z. Y. Huang, D. R. Green, D. R. Smith and J. Zhang,
Cline, T. W., and B. J. Meyer,
Cook, J.,
Cristino, A. S., A. M. Nascimento, F. Costa Lda and Z. L. Simoes,
Erdman, S. E., and K. C. Burtis,
Funaguma, S., M. G. Suzuki, T. Tamura and T. Shimada,
Graham, P., J. K. Penn and P. Schedl,
Hasselmann, M., and M. Beye,
Hediger, M., G. Burghardt, C. Siegenthaler, N. Buser, D. Hilfiker-Kleiner et al.,
Honeybee Genome Sequencing Consortium,
Inoue, K., K. Hoshijima, I. Higuchi, H. Sakamoto and Y. Shimura,
Kuhn, S., V. Sievert and W. Traut,
Lagos, D., M. F. Ruiz, L. Sanchez and K. Komitopoulou,
Lagos, D., M. Koukidou, C. Savakis and K. Komitopoulou,
Lynch, K. W., and T. Maniatis,
Marin, I., and B. S. Baker,
Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda et al.,
Meise, M., D. Hilfiker-Kleiner, A. Dubendorfer, C. Brunner, R. Nothiger et al.,
Nanda, I., M. Kondo, U. Hornung, S. Asakawa, C. Winkler et al.,
Niimi, T., K. Sahara, H. Oshima, Y. Yasukochi, K. Ikeo et al.,
Ohbayashi, F., M. G. Suzuki, K. Mita, K. Okano and T. Shimada,
Pomiankowski, A., R. Nothiger and A. Wilkins,
Pane, A., M. Salvemini, P. Delli Bovi, C. Polito and G. Saccone,
Pane, A., A. De Simone, G. Saccone and C. Polito,
Raymond, C. S., C. E. Shamu, M. M. Shen, K. J. Seifert, B. Hirsch et al.,
Raymond, C. S., M. W. Murphy, M. G. O'Sullivan, V. J. Bardwell and D. Zarkower,
Ruiz, M. F., R. N. Stefani, R. O. Mascarenhas, A. L. Perondini, D. Selivon et al.,
Saccone, G., I. Peluso, D. Artiaco, E. Giordano, D. Bopp et al.,
Savard, J., D. Tautz, S. Richards, G. M. Weinstock, R. A. Gibbs et al.,
Scali, C., F. Catteruccia, Q. Li and A. Crisanti,
Shearman, D. C., and M. Frommer,
Sievert, V., S. Kuhn, A. Paululat and W. Traut,
Suzuki, M. G., F. Ohbayashi, K. Mita and T. Shimada,
Suzuki, M. G., S. Funaguma, T. Kanda, T. Tamura and T. Shimada,
Suzuki, M. G., S. Funaguma, T. Kanda, T. Tamura and T. Shimada,
Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins,
Traut, W., T. Niimi, K. Ikeo and K. Sahara,
Wilkins, A. S.,
Zhang, J.,