-
PDF
- Split View
-
Views
-
Cite
Cite
Marna D Yandeau-Nelson, Yiji Xia, Jin Li, M Gerald Neuffer, Patrick S Schnable, Unequal Sister Chromatid and Homolog Recombination at a Tandem Duplication of the a1 Locus in Maize, Genetics, Volume 173, Issue 4, 1 August 2006, Pages 2211–2226, https://doi.org/10.1534/genetics.105.052712
Close - Share Icon Share
Abstract
Tandemly arrayed duplicate genes are prevalent. The maize A1-b haplotype is a tandem duplication that consists of the components, α and β. The rate of meiotic unequal recombination at A1-b is ninefold higher when a homolog is present than when it is absent (i.e., hemizygote). When a sequence heterologous homolog is available, 94% of recombinants (264/281) are generated via recombination with the homolog rather than with the sister chromatid. In addition, 83% (220/264) of homolog recombination events involved α rather than β. These results indicate that: (1) the homolog is the preferred template for unequal recombination and (2) pairing of the duplicated segments with the homolog does not occur randomly but instead favors a particular configuration. The choice of recombination template (i.e., homolog vs. sister chromatid) affects the distribution of recombination breakpoints within a1. Rates of unequal recombination at A1-b are similar to the rate of recombination between nonduplicated a1 alleles. Unequal recombination is therefore common and is likely to be responsible for the generation of genetic variability, even within inbred lines.
TANDEMLY arrayed duplicate genes are prevalent across species. As defined by Zhang and Gaut (2003), tandemly arrayed duplicate genes are paralogs that are physically separated on a chromosome by 0–10 unrelated “spacer genes.” In Caenorhabditis elegans, 10% of mapped genes are tandem duplicates (Semple and Wolfe 1999). At similar frequencies, tandemly arrayed duplicate genes are present in plant genomes, including within Arabidopsis (∼12–16%; Arabidopsis Genome Initiative 2000; Zhang and Gaut 2003) and rice (14%; International Rice Genome Sequencing Project 2005). Within maize an even higher percentage of genes (∼33%) are estimated to be members of tandem arrays (Messing et al. 2004). By virtue of their high level of shared sequence identity, paralogs within a tandemly arrayed duplicate gene family can misalign and pair unequally in meiosis. Unequal recombination between these unequally paired paralogs, or components, within a duplication can occur via the double-strand break repair (DSBR) model (Szostak et al. 1983; Sun et al. 1991; Allers and Lichten 2001) of recombination.
Unequal recombination between tandemly arrayed duplicate genes was first documented at the bar locus in Drosophila (Sturtevant 1925). Since then, the process has been proposed as the mechanism for the evolution of many tandemly arrayed duplicate gene families (reviewed in Zhang 2003), including the HOX genes (reviewed in Garcia-Fernandez 2005), ribosomal DNA repeats (Williams and Robbins 1992; reviewed in Petes and Pukkila 1995), and plant resistance genes (reviewed in Leister 2004) in lettuce (Chin et al. 2001; Kuang et al. 2004) and in maize including the rp1 (Sudupak et al. 1993; Richter et al. 1995; Sun et al. 2001; Ramakrishna et al. 2002; Smith and Hulbert 2005) and rp3 (Webb et al. 2002) gene clusters. Unequal recombination between tandemly duplicated genes can generate novel alleles as has been observed in rp1 (Sun et al. 2001), rp3 (Webb et al. 2002), R-r (Stadler and Nuffer 1953; Dooner and Kermicle 1971, 1974; Robbins et al. 1991; Walker et al. 1995), bz1 (Dooner and Martinez-Ferez 1997), kn1 (Lowe et al. 1992), the 27-kDa zein (Das et al. 1991), and p1 (Zhang and Peterson 2005).
Recombination, either between duplicate or single-copy genes, can occur between both sister chromatids (i.e., interchromatid recombination) and homologs (i.e., interhomolog recombination). At single-copy genes in yeast and mammals, mitotic reciprocal recombination occurs preferentially between sister chromatids (Kadyk and Hartwell 1992; reviewed in Johnson and Jasin 2001; Gonzalez-Barrera et al. 2003) but with the onset of meiosis the preferred recombination template changes to the homolog (reviewed in Petes and Pukkila 1995; Kleckner 1996; Roeder 1997). Similarly in yeast, unequal recombination between duplicated sequences occurs primarily between sister chromatids during mitosis (Jackson and Fink 1981), whereas during meiosis, unequal interhomolog recombination occurs 10-fold more frequently than interchromatid recombination (Jackson and Fink 1985).
In plants, unequal recombination occurs at the naturally occurring maize R-r and A1-b tandem duplications at rates ranging between 10−3 and 10−4 (Laughnan 1952; Dooner and Kermicle 1971). In the absence of molecular characterization it was not possible to distinguish between interhomolog and interchromatid recombination. Even so, the findings that (1) rates of unequal recombination at A1-b were higher in the presence of a homolog than in its absence (i.e., hemizygotes) and (2) recombinants isolated from marked heterozygotes most often showed an exchange of flanking phenotypic markers (Laughnan 1949, 1952), were interpreted to suggest that the homolog is the preferred recombination template (Laughnan 1952). More recently, molecular characterizations in plants have documented both meiotic interhomolog (Assaad and Signer 1992; Tovar and Lichtenstein 1992; Molinier et al. 2004) and interchromatid (Assaad and Signer 1992; Tovar and Lichtenstein 1992; Jelesko et al. 1999, 2004; Molinier et al. 2004) unequal recombination events. Meiotic unequal recombination events isolated from synthetic direct repeats in Arabidopsis (Molinier et al. 2004) and synthetic inverted repeats in tobacco (Tovar and Lichtenstein 1992) were conducted using both homozygotes (in which recombination can occur either between homologs or between sister chromatids) and hemizygotes (in which recombination can occur only between sister chromatids). Although the recombination template (i.e., homolog vs. sister chromatid) cannot be directly determined in homozygotes, the finding that unequal recombination occurred more than two times as frequently in the homozygotes than in hemizygotes has been interpreted to suggest that in plants meiotic unequal interhomolog recombination occurs more often than interchromatid recombination.
Although several studies in plants (Assaad and Signer 1992; Tovar and Lichtenstein 1992; Molinier et al. 2004) have identified the mechanism by which unequal recombinants were generated (e.g., gene conversion and interchromosomal or intrachromosomal unequal recombination), the recombination breakpoints associated with unequal recombination events have been analyzed in only two related studies. Unequal recombination breakpoints (n = 25) were distributed nonrandomly and were correlated with regions of higher sequence identity in recombinants isolated from hemizygotes (i.e., unequal recombination between sister chromatids) in a synthetic tandemly arrayed cluster of RBCSB genes (Jelesko et al. 1999, 2004).
Several characteristics of unequal recombination between the components of gene duplications remain to be resolved in plants and other organisms. These include defining directly the frequencies of interhomolog and interchromatid unequal recombination, how rates and patterns of meiotic unequal interhomolog and interchromatid recombination are differently regulated, and which additional factors might affect rates and/or patterns of unequal recombination. In addition, it is not known whether all possible pairings of components of a duplication with the homolog (i.e., the pairing configuration) occur at equal frequencies. Finally, extant analyses of unequal recombination between synthetic repeat constructs do not allow for comparisons of rates or patterns of recombination with corresponding single-copy alleles in the same genomic location. In plants this is because, absent homologous gene replacement, it is not possible to obtain a stock that contains a synthetic duplication at the same genomic location as a related single-copy sequence.
The A1-b tandem duplication of the maize a1 gene is an ideal system in which to address these unanswered questions regarding unequal recombination because: (1) A1-b is a naturally occurring tandem duplication and genetic stocks containing single-copy a1 alleles at the same genomic location are available, (2) a large portion of the ∼140-kb a1-sh2 interval on chromosome 3L has been sequenced and PCR-based genetic markers across the interval are available (Xu et al. 1995; Yao et al. 2002), and (3) unequal recombination events across the interval can be easily identified by their nonparental kernel phenotypes.
This study extends earlier studies of the A1-b tandem gene duplication conducted by Laughnan (1949, 1952, 1955) by defining the molecular structure of the duplication and directly demonstrating that unequal recombination in the A1-b tandem duplication occurs preferentially, and at very high rates, between homologs. In addition, this study establishes that the choice of recombination template (homolog vs. sister chromatid) significantly affects the distribution of recombination breakpoints. Further, the choice of unequal pairing configuration of the duplicated components with the homolog is not random. The similar rates of unequal recombination between the components of A1-b and equal recombination between nonduplicated a1 alleles suggest that unequal recombination is common and likely contributes to genetic variability, even within genetic stocks and inbred lines in maize.
MATERIALS AND METHODS
The a1-sh2 interval:
The ∼140-kb a1-sh2 interval (Civardi et al. 1994) contains four genes, a1, yz1, x1, and sh2 (Figure 1A; Yao et al. 2002). Mutations in the a1 and sh2 loci condition colorless aleurone and shrunken kernel phenotypes, respectively. A Ds transposon insertion in exon 2 of yz1 does not exhibit any obvious mutant phenotype (data not shown).
Physical characterization of deletion alleles at a1. (A) Structure of single-component A1-Sh2 haplotypes (Yao et al. 2002). Boxes represent genes. IR, interloop region (Yao et al. 2002). (B) Physical characterization of deletion alleles at a1. DNA gel blot analyses of putative a1-sh2 deletion stocks are shown. Genomic DNAs were digested with HindIII and hybridized with an a1-specific probe (materials and methods).
Alleles and genetic stocks:
A1-b and A1-b(P415) (obtained from the Maize Genetics Stock Center; Co-op ID 315D), which are equivalent to Ab and Ab: P (Laughnan 1949, 1952, 1955), are both tandem duplications of the a1 locus consisting of α and β components. In A1-b and A1-b(P415), the orders of components are centromere-α-β-Sh2 (Laughnan 1949) and centromere-β-α-Sh2 (Laughnan 1955), respectively. The α and β components confer pale and colored aleurones, respectively, and, with respect to aleurone phenotype, β is dominant to α. The a1∷rdt sh2 haplotype was described previously (reviewed in Xu et al. 1995) and confers colorless shrunken kernels. Kernels homozygous for the a1∷rdt Sh2 haplotype are colorless and round. The a1-s (also designated as a1-dl) allele was described by Hsia and Schnable (1996) and the a1-mum2 allele by Schwarz-Sommer et al. (1987) and Xu et al. (1995). The A1′-91g103 sh2 haplotype was generated via an intragenic recombination event isolated from an a1-mum2 Sh2/a1∷rdt sh2 heterozygote (data not shown). The a1-m3 Sh2 haplotype and the compound a1 alleles α A1 Sh2 and α a1-m3 Sh2 were described by Neuffer (1965) and obtained from M. Gerald Neuffer.
EMS-induced point mutant alleles of a1, which condition a colorless phenotype, were generated and confirmed as described in Hsia et al. (2005). The a1-3849-9 Sh2 et (GenBank accession no. DQ017580) and a1-3849-5 Sh2 et (GenBank accession no. DQ012666) haplotypes were derived from the A1-Cornell (GenBank accession no. U46055) allele and contain a C-to-T transition that changed a glutamine to a stop codon in exon 4 of a1 (position 2891 of GenBank accession no. X05068) and a C-to-T transition that changed a phenylalanine to a serine in exon 3 (position 798 of GenBank accession no. U46055), respectively. In the a1-3845-5 Sh2 Et haplotype (GenBank accession no. DQ012661) derived from A1-Line C (GenBank accession no. X0506), which is derived from a color-converted version of the W22 inbred, a C-to-T transition changed a phenylalanine to a serine in exon 3 (position 2438 of GenBank accession no. X05068). The et1 gene encodes a plastid-specific transcription factor (da Costa e Silva et al. 2004) and is ∼10 cM distal to Sh2. Kernels homozygous for the recessive et1 allele exhibit a cracked surface (Stadler 1940).
Characterization of stocks deficient for the a1-sh2 interval:
The ax-1, ax-2, and ax-3 deficiency stocks identified from populations derived from X-ray-treated pollen by virtue of their colorless aleurone phenotypes were considered to be deficient for a1 and surrounding sequences on chromosome 3L (Stadler and Roman 1948). The del stock is also a putative deletion of the a1-sh2 interval identified from progeny of an a1-mum2 allele in a Mu-active background (Stinard and Robertson 1988). Likewise, the df genetic stock provided by M. G. Neuffer is putatively deficient for the a1-sh2 interval. Because these putative a1-sh2 deficiencies are homozygous lethal, each was maintained as a heterozygote with either A1′-91g103 sh2 or a1-mum2 sh2. DNA gel blot analyses (Sambrook et al. 1989) were conducted using HindIII-digested genomic DNA isolated (Dellaporta et al. 1983) from each deficiency heterozygote (with A1′-91g103 sh2 or a1-mum2 sh2). The a1 probe was PCR amplified from the a1 genomic clone pA1 (Civardi et al. 1994) with primers QZ1003 (5′ ATA ATA GTA GCC TCC CGA ATA A 3′) and A1522 (Table 1); the yz1 probe was PCR amplified from the cDNA clone yz1.hy1 (Yao et al. 2005) using primers yz3UTRF (Table 1) and YZC7 (5′ CGAAGCCACCGCAAGC 3′); the x1 probe was PCR amplified from the cDNA clone X-V1 (Yao et al. 2002) using primers X501 (5′ CGAGGCAAAAGAAAAAGCAGT 3′) and X301 (5′ CTTATCGCTTCCTCCTGTTTG 3′); and the sh2 probe was PCR amplified from the cDNA clone pcSh2-1a (Bhave et al. 1990) using primers sh2c205 (5′ CTTTGAGAAATAGGTGCTTTGG 3′) and sh2c853r (5′ AAAGAATTGAAGTACACGTCCAG 3′).
Oligonucleotides used for PCR-based recombinant mapping
. | . | . | a1 haplotype . | ||
|---|---|---|---|---|---|
| Primer name . | Sequencea . | Polymorphism . | α-Yz1Ab . | β-Sh2c . | a1∷rdt sh2 . |
| αSNP4R | GTGTGGGGTCTAGAGAAGGG | SNP | +d | −e | − |
| αSNP5R | GCTTGAGGATCGAGTAGTGC | SNP | + | − | − |
| αIDP1R | TGAGAAACTTCTTTCGGCTCTG | IDP | + | − | − |
| αSNP6R | CAACACCAAACCCTTCAACCA | SNP | + | − | − |
| αSNP3R | CCAGCCTTTTTATCCCGCTC | SNP | + | − | − |
| αSNP3F | GCAAGAACACATTAGACACGTTA | SNP | + | − | − |
| β3694R | GTCTTCCCCACATAATATGCG | SNP | − | + | − |
| β3040R | CGAGGAGCAGACGTAGCGG | SNP | − | + | − |
| β2149R | CAACGTTCGCTGCAGGAC | SNP | − | + | − |
| yz4725 | AAATGGTCAGGATAGCTTAGTT | IDP | + | + | |
| yz5U-AbR | GGCTCCATATATCAAGCACA | IDP | + | + | − |
| Sh5379R | ACCAATGATACAGAGAGGCG | SNP | NAf | + | − |
| rdt444 | AGCAAATAGCAATAATCAAGGCA | IDPg | − | − | + |
| rdtILR1 | AGACAAATGTTCTGTAGGAAAC | SNP | − | − | + |
| yzrdtIDP1 | GTTCACACAAAGTATTTTTTTCG | IDP | − | − | + |
| yz1410R | GGCTCCATATATCAAGCAGT | IDP | − | − | + |
| Shrdt1R | GACCAATGATACAGAGAGGCA | SNP | − | − | + |
| QZ1504 | CCAGGGGATAAAACAATTCGT | U | + | + | + |
| A2775F | CACCATCATCCCGACGCTC | U | + | + | + |
| A2357 | AGCCGACGGTGGAAGGGATG | U | + | + | + |
| XX390h | TCGGCTTGATTACCTCATTCT | U | + | + | + |
| A6458f | GGGAAGACGAAGCCATTGA | U | + | + | + |
| QZ1265h | TACTCCTCTCCAACTCCA | U | + | + | + |
| A1522 | GGGAGTTTGGAGTTGGAGAGG | U | + | + | + |
| ajl002 | TCAAGCTAAAAGAAAGAAACATT | U | + | + | + |
| a178R | TGCCAAATAACCATACCACA | U | + | + | + |
| QZ1742 | TAGTTGGTAGCACGGTTGA | U | + | + | + |
| yz3UTRF | CGGGGGTTGCAGTCATTGAC | U | + | + | + |
| ZH1748 | CACATCCCCGTCTCCT | U | + | + | + |
| yz792F | GCGGTTGCGGCTTGTAC | U | + | + | + |
| MY339 | GCCTTTCCCCCATTACTATC | U | + | + | + |
. | . | . | a1 haplotype . | ||
|---|---|---|---|---|---|
| Primer name . | Sequencea . | Polymorphism . | α-Yz1Ab . | β-Sh2c . | a1∷rdt sh2 . |
| αSNP4R | GTGTGGGGTCTAGAGAAGGG | SNP | +d | −e | − |
| αSNP5R | GCTTGAGGATCGAGTAGTGC | SNP | + | − | − |
| αIDP1R | TGAGAAACTTCTTTCGGCTCTG | IDP | + | − | − |
| αSNP6R | CAACACCAAACCCTTCAACCA | SNP | + | − | − |
| αSNP3R | CCAGCCTTTTTATCCCGCTC | SNP | + | − | − |
| αSNP3F | GCAAGAACACATTAGACACGTTA | SNP | + | − | − |
| β3694R | GTCTTCCCCACATAATATGCG | SNP | − | + | − |
| β3040R | CGAGGAGCAGACGTAGCGG | SNP | − | + | − |
| β2149R | CAACGTTCGCTGCAGGAC | SNP | − | + | − |
| yz4725 | AAATGGTCAGGATAGCTTAGTT | IDP | + | + | |
| yz5U-AbR | GGCTCCATATATCAAGCACA | IDP | + | + | − |
| Sh5379R | ACCAATGATACAGAGAGGCG | SNP | NAf | + | − |
| rdt444 | AGCAAATAGCAATAATCAAGGCA | IDPg | − | − | + |
| rdtILR1 | AGACAAATGTTCTGTAGGAAAC | SNP | − | − | + |
| yzrdtIDP1 | GTTCACACAAAGTATTTTTTTCG | IDP | − | − | + |
| yz1410R | GGCTCCATATATCAAGCAGT | IDP | − | − | + |
| Shrdt1R | GACCAATGATACAGAGAGGCA | SNP | − | − | + |
| QZ1504 | CCAGGGGATAAAACAATTCGT | U | + | + | + |
| A2775F | CACCATCATCCCGACGCTC | U | + | + | + |
| A2357 | AGCCGACGGTGGAAGGGATG | U | + | + | + |
| XX390h | TCGGCTTGATTACCTCATTCT | U | + | + | + |
| A6458f | GGGAAGACGAAGCCATTGA | U | + | + | + |
| QZ1265h | TACTCCTCTCCAACTCCA | U | + | + | + |
| A1522 | GGGAGTTTGGAGTTGGAGAGG | U | + | + | + |
| ajl002 | TCAAGCTAAAAGAAAGAAACATT | U | + | + | + |
| a178R | TGCCAAATAACCATACCACA | U | + | + | + |
| QZ1742 | TAGTTGGTAGCACGGTTGA | U | + | + | + |
| yz3UTRF | CGGGGGTTGCAGTCATTGAC | U | + | + | + |
| ZH1748 | CACATCCCCGTCTCCT | U | + | + | + |
| yz792F | GCGGTTGCGGCTTGTAC | U | + | + | + |
| MY339 | GCCTTTCCCCCATTACTATC | U | + | + | + |
IDP, insertion/deletion polymorphism; SNP, single-nucleotide polymorphism; U, universal primer.
Sequences are listed 5′–3′.
Sequences specific to the interval in A1-b Sh2 extending from α to Yz1A.
Sequences specific to the interval in A1-b Sh2 extending from β to Sh2.
+ indicates a primer that can amplify the corresponding interval.
− indicates a primer that cannot amplify the corresponding interval.
Sh2 is not within the α-Yz1A interval. NA, not applicable.
Primer specific to the rdt transposon insertion.
Both primers (XX390 and QZ1265) amplify in each of the three haplotypes. In combination, however, these primers amplify a product only in the α-Yz1 interval.
Oligonucleotides used for PCR-based recombinant mapping
. | . | . | a1 haplotype . | ||
|---|---|---|---|---|---|
| Primer name . | Sequencea . | Polymorphism . | α-Yz1Ab . | β-Sh2c . | a1∷rdt sh2 . |
| αSNP4R | GTGTGGGGTCTAGAGAAGGG | SNP | +d | −e | − |
| αSNP5R | GCTTGAGGATCGAGTAGTGC | SNP | + | − | − |
| αIDP1R | TGAGAAACTTCTTTCGGCTCTG | IDP | + | − | − |
| αSNP6R | CAACACCAAACCCTTCAACCA | SNP | + | − | − |
| αSNP3R | CCAGCCTTTTTATCCCGCTC | SNP | + | − | − |
| αSNP3F | GCAAGAACACATTAGACACGTTA | SNP | + | − | − |
| β3694R | GTCTTCCCCACATAATATGCG | SNP | − | + | − |
| β3040R | CGAGGAGCAGACGTAGCGG | SNP | − | + | − |
| β2149R | CAACGTTCGCTGCAGGAC | SNP | − | + | − |
| yz4725 | AAATGGTCAGGATAGCTTAGTT | IDP | + | + | |
| yz5U-AbR | GGCTCCATATATCAAGCACA | IDP | + | + | − |
| Sh5379R | ACCAATGATACAGAGAGGCG | SNP | NAf | + | − |
| rdt444 | AGCAAATAGCAATAATCAAGGCA | IDPg | − | − | + |
| rdtILR1 | AGACAAATGTTCTGTAGGAAAC | SNP | − | − | + |
| yzrdtIDP1 | GTTCACACAAAGTATTTTTTTCG | IDP | − | − | + |
| yz1410R | GGCTCCATATATCAAGCAGT | IDP | − | − | + |
| Shrdt1R | GACCAATGATACAGAGAGGCA | SNP | − | − | + |
| QZ1504 | CCAGGGGATAAAACAATTCGT | U | + | + | + |
| A2775F | CACCATCATCCCGACGCTC | U | + | + | + |
| A2357 | AGCCGACGGTGGAAGGGATG | U | + | + | + |
| XX390h | TCGGCTTGATTACCTCATTCT | U | + | + | + |
| A6458f | GGGAAGACGAAGCCATTGA | U | + | + | + |
| QZ1265h | TACTCCTCTCCAACTCCA | U | + | + | + |
| A1522 | GGGAGTTTGGAGTTGGAGAGG | U | + | + | + |
| ajl002 | TCAAGCTAAAAGAAAGAAACATT | U | + | + | + |
| a178R | TGCCAAATAACCATACCACA | U | + | + | + |
| QZ1742 | TAGTTGGTAGCACGGTTGA | U | + | + | + |
| yz3UTRF | CGGGGGTTGCAGTCATTGAC | U | + | + | + |
| ZH1748 | CACATCCCCGTCTCCT | U | + | + | + |
| yz792F | GCGGTTGCGGCTTGTAC | U | + | + | + |
| MY339 | GCCTTTCCCCCATTACTATC | U | + | + | + |
. | . | . | a1 haplotype . | ||
|---|---|---|---|---|---|
| Primer name . | Sequencea . | Polymorphism . | α-Yz1Ab . | β-Sh2c . | a1∷rdt sh2 . |
| αSNP4R | GTGTGGGGTCTAGAGAAGGG | SNP | +d | −e | − |
| αSNP5R | GCTTGAGGATCGAGTAGTGC | SNP | + | − | − |
| αIDP1R | TGAGAAACTTCTTTCGGCTCTG | IDP | + | − | − |
| αSNP6R | CAACACCAAACCCTTCAACCA | SNP | + | − | − |
| αSNP3R | CCAGCCTTTTTATCCCGCTC | SNP | + | − | − |
| αSNP3F | GCAAGAACACATTAGACACGTTA | SNP | + | − | − |
| β3694R | GTCTTCCCCACATAATATGCG | SNP | − | + | − |
| β3040R | CGAGGAGCAGACGTAGCGG | SNP | − | + | − |
| β2149R | CAACGTTCGCTGCAGGAC | SNP | − | + | − |
| yz4725 | AAATGGTCAGGATAGCTTAGTT | IDP | + | + | |
| yz5U-AbR | GGCTCCATATATCAAGCACA | IDP | + | + | − |
| Sh5379R | ACCAATGATACAGAGAGGCG | SNP | NAf | + | − |
| rdt444 | AGCAAATAGCAATAATCAAGGCA | IDPg | − | − | + |
| rdtILR1 | AGACAAATGTTCTGTAGGAAAC | SNP | − | − | + |
| yzrdtIDP1 | GTTCACACAAAGTATTTTTTTCG | IDP | − | − | + |
| yz1410R | GGCTCCATATATCAAGCAGT | IDP | − | − | + |
| Shrdt1R | GACCAATGATACAGAGAGGCA | SNP | − | − | + |
| QZ1504 | CCAGGGGATAAAACAATTCGT | U | + | + | + |
| A2775F | CACCATCATCCCGACGCTC | U | + | + | + |
| A2357 | AGCCGACGGTGGAAGGGATG | U | + | + | + |
| XX390h | TCGGCTTGATTACCTCATTCT | U | + | + | + |
| A6458f | GGGAAGACGAAGCCATTGA | U | + | + | + |
| QZ1265h | TACTCCTCTCCAACTCCA | U | + | + | + |
| A1522 | GGGAGTTTGGAGTTGGAGAGG | U | + | + | + |
| ajl002 | TCAAGCTAAAAGAAAGAAACATT | U | + | + | + |
| a178R | TGCCAAATAACCATACCACA | U | + | + | + |
| QZ1742 | TAGTTGGTAGCACGGTTGA | U | + | + | + |
| yz3UTRF | CGGGGGTTGCAGTCATTGAC | U | + | + | + |
| ZH1748 | CACATCCCCGTCTCCT | U | + | + | + |
| yz792F | GCGGTTGCGGCTTGTAC | U | + | + | + |
| MY339 | GCCTTTCCCCCATTACTATC | U | + | + | + |
IDP, insertion/deletion polymorphism; SNP, single-nucleotide polymorphism; U, universal primer.
Sequences are listed 5′–3′.
Sequences specific to the interval in A1-b Sh2 extending from α to Yz1A.
Sequences specific to the interval in A1-b Sh2 extending from β to Sh2.
+ indicates a primer that can amplify the corresponding interval.
− indicates a primer that cannot amplify the corresponding interval.
Sh2 is not within the α-Yz1A interval. NA, not applicable.
Primer specific to the rdt transposon insertion.
Both primers (XX390 and QZ1265) amplify in each of the three haplotypes. In combination, however, these primers amplify a product only in the α-Yz1 interval.
For each of the four genes within the a1-sh2 interval, an RFLP exists between the A1′-91g103 sh2 and a1-mum2 sh2 haplotypes (Figure 1B). If any of the putative deficiencies carries a copy of the a1 gene, a second DNA fragment would be expected to hybridize to the a1 probe in either or both of the A1′-91g103 sh2 and a1-mum2 sh2 deletion heterozygotes. Instead, the a1 gene probe detects only a single fragment in each of these heterozygotes (Figure 1B), demonstrating that a1 is deleted in ax-1, ax-2, ax-3, df, and del. A similar strategy was applied to the yz1, x1, and sh2 genes with similar results, demonstrating that the entire a1-sh2 interval is absent in each of the five deficiencies (data not shown).
Cloning of A1-b α and β:
Genomic DNA isolated from A1-b/A1-b plants (Schnable Lab pedigree no. 93-4216-21 self) was digested with HindIII and subjected to DNA gel blot analysis (Sambrook et al. 1989). A 0.6-kb a1 probe corresponding to exons 2–3 of a1 generated by PstI digestion of pCU9 hybridized to 18-kb and 5.8-kb HindIII fragments (Figure 2, lane 4). Similar DNA gel blot analyses were conducted with stocks containing compound a1 alleles in which the β-component had been replaced by another a1 allele (Figure 2, lanes 2, 3, and 5) via recombination between A1-b and other a1 alleles (Neuffer 1965). The 18-kb but not the 5.8-kb fragment was detectable in the compound stocks containing α but not β (Figure 2, lanes 2, 3, and 5), demonstrating that the 18-kb A1-b fragment includes the α-component. HindIII-digested DNA fragments of 15–20 kb and 5–7 kb were cloned into the λ-insertion vector NM1149 and replacement vector λDashII, respectively, and packaged. Three a1-hybridizing clones were analyzed from each library.
Physical identification of the α- and β-components of the A1-b allele. Homozygous genomic DNAs derived from a single-component a1 haplotype, a1-m3 Sh2, and double-component a1 haplotypes, α A1 Sh2, α a1-m3 Sh2, A1-b Sh2, and α A1 Sh2, were digested with HindIII and hybridized with an a1 probe (materials and methods). All four double-component a1 haplotypes contain the α-component (∼18 kb) but only A1-b Sh2 contains the β-component (5.8 kb). The two double-component a1 haplotypes designated α A1 Sh2 contain different a1 alleles.
Sequencing of A1-b and A1-b(P415):
Sequences of α (Figure 3B) and β from the A1-b haplotype (GenBank accession nos. DQ219416 and DQ219417) were obtained by primer walking across subclones and sequencing PCR fragments derived from the α-containing λA-b-α-2 clone and the β-containing pAb-β-1 clone as described in supplemental data (http://www.genetics.org/supplemental/). Partial sequences of α and β from the A1-b(P415) haplotype (GenBank accession nos. DQ219418 and DQ219419) were obtained from PCR fragments derived from the A1-b(P415) genetic stock as described in supplemental data. For all sequences, both strands were sequenced. Sequences were analyzed and assembled using Sequencher software (version 4.2.2; Gene Codes, Ann Arbor, MI).
Structure of the A1-b Sh2 haplotype. (A) The A1-b Sh2 haplotype. The sequence identities and sizes of duplicated segments are shown. Black lines designate regions with 100% sequence identity. Boxes and lines designate genes and intergenic regions, respectively. It is not known whether the x1 gene [which is ∼35 kb distal to yz1 in nonduplicated haplotypes (Yao et al. 2002)] is included in the duplication associated with the A1-b haplotype. Not drawn to scale. IR, interloop region (Yao et al. 2002). (B) Structure of A1-b α. A 5.4-kb insertion containing a 3.8-kb transposon-like element that contains a nested 453-bp Ins2 element with 93% identity to Ins2 in bz-R (GenBank accession no. X07938) is located within intron 2. The 3.8-kb element is flanked by 458-bp terminal inverted repeats (TIRs; long thick arrows), for which the first 13 bp of the TIRs are identical to the 13-bp TIRs of Ins2 (short arrows). The distal 458-bp TIR is actually a part of an intact 636-bp transposable element (triangle) with 93% identity to an uncharacterized transposon in intron 3 of the maize fatty aldehyde dehydrogenase1 gene (GenBank accession no. AY374447). Directly adjacent to the 3.8-kb element is a 1.6-kb inverted duplication of a1 sequence (shown in boldface type). Flanking the 5.4-kb insertion are 21-bp flanking direct duplications (solid bars). A Cin4 retrotransposon insertion is located in exon 4 at the same position as within other type II A1 alleles (Schwarz-Sommer et al. 1987) but is not present in the A1-b β-component. Boxes and lines represent exons and introns, respectively. Triangles represent insertions and arrows identify TIRs.
Isolation and confirmation of unequal recombination events at A1-b:
Recombinants were isolated from tandem duplication homozygotes (Figure 4A) and from tandem duplication heterozygotes in which both interhomolog and interchromatid recombination can occur (Figure 4B). Because the interhomolog and interchromatid recombinant gametes differ in structure (Figure 4B), the rates of interhomolog and interchromatid recombination can be directly and separately measured in this genotype.
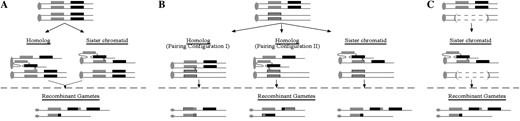
Strategy for isolating and distinguishing interhomolog and interchromatid recombinants. (A) Isolation of unequal recombinants from tandem duplication homozygotes. Unequal pairing configurations between the sequence-identical homologs and sister chromatids within a homozygote yield recombinant gametes with identical structures and therefore make it impossible to determine which recombination template was used. (B) Isolation of unequal recombinants from tandem duplication heterozygotes. Recombination in a heterozygote with a homolog containing a tandem gene duplication and a homolog containing a corresponding sequence heterologous single-copy gene can be detected between homologs and sister chromatids by virtue of the molecularly distinguishable recombinant structures. Because the homolog containing the single-copy gene can pair with either component of the tandem duplication (resulting in two distinct pairing configurations), the relative frequencies of alternative pairing configurations with the homolog can also be measured. (C) Isolation of unequal recombinants from tandem duplication hemizygotes. Because the duplicated locus is present only on one homolog in a hemizygote, recombinants isolated from a hemizygote must have occurred via interchromatid recombination. (A–C) The shaded and solid rectangles represent the components of a tandem duplication; (B) the hatched rectangle represents a paralog that is heterologous in sequence as compared to the components of the tandem duplication. Circles and ovals designate centromeres and an “x” designates the position of recombination.
To identify recombination events at A1-b both in the presence and in the absence of an a1 homolog, the A1-b Sh2 genetic stock was crossed as male to a1∷rdt Sh2/ax-1 heterozygotes (cross 1A). Recombination in the A1-b Sh2/a1∷rdt Sh2 F1's resulting from cross 1A (cross 2A) can occur between either homologs (Figure 5, classes III and IV) or sister chromatids (Figure 5, class V). Because the other progeny of cross 1A used as the female parent of cross 3A are hemizygous for A1-b Sh2 (the a1-sh2 interval is deleted from the ax-1 haplotype, Figure 1B) recombination can occur only between sister chromatids (similar to that depicted in Figure 5, class V). Recombination was measured in female parents of crosses 2A and 3A and in A1-b homozygotes (cross 4A) according to previously described methods for the isolation and confirmation of recombinants across the a1-sh2 interval (Xu et al. 1995; Yao and Schnable 2005). Crosses similar to those provided in crosses 1A–4A were also conducted using the A1-b(P415) haplotype (crosses 1B–4B):
Cross 1A: a1∷rdt Sh2/ax-1 x A1-b Sh2/A1-b Sh2
Cross 1B: a1∷rdt Sh2/ax-1 x A1-b(P415) Sh2/A1-b(P415) Sh2
Cross 2A: A1-b Sh2/a1∷rdt Sh2 x a1-s sh2/a1-s sh2 or a1∷rdt sh2/a1∷rdt sh2
Cross 2B: A1-b(P415) Sh2/a1∷rdt Sh2 x a1∷rdt sh2/a1∷rdt sh2
Cross 3A: A1-b Sh2/ax-1 x a1-s sh2/a1-s sh2 or a1∷rdt sh2/a1∷rdt sh2
Cross 3B: A1-b(P415) Sh2/ax-1 x a1∷rdt sh2/a1∷rdt sh2
Cross 4A: A1-b Sh2/A1-b Sh2 x a1-s sh2/a1-s sh2 or a1∷rdt sh2/a1∷rdt sh2
Cross 4B: A1-b(P415) Sh2/A1-b(P415) Sh2 x a1∷rdt sh2/a1∷rdt sh2
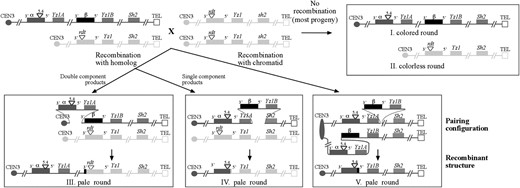
Isolation of recombinants. The parents of and the progeny resulting from cross 2A (materials and methods) are illustrated. Chromosomes from the A1-b Sh2 and a1∷rdt Sh2 stocks are illustrated as solid and shaded, respectively. Triangles indicate the positions of the 5.4-kb insertion in α and the rdt transposon in the single-copy a1 allele. Circles (or ovals) and squares represent centromeres and telomeres, respectively. Although all recombination breakpoints are illustrated as resolving within a1, resolution could potentially occur anywhere in the region between the 5.4-kb insertion of α and the β-component. In classes IV and V, Yz1A is illustrated as pairing with either Yz1 on the homolog (class IV) or Yz1B on the sister chromatid (class V); however, alternative pairing configurations in which Yz1B pairs with the homolog (class IV) or sister chromatid (class V) are also possible. Recombination between equally paired sister chromatids would not be detected in this assay. Unequal interchromatid recombinants from cross 3A would resemble the class V recombinants illustrated here. Unequal interhomolog and interchromatid recombinants from A1-b homozygotes (cross 4A) cannot be distinguished and would both resemble the class V recombinants illustrated here. In crosses 2–4, recombination might also occur between the inverted duplicate sequences within α (Figure 3B). Such recombination events would generate acentric or dicentric products, which would not be recovered following meiosis.
The vast majority of resulting progeny kernels were of parental phenotype in cross 2 (viz., colored and colorless round), cross 3 (viz., colored round and colorless shrunken), and cross 4 (viz., colored round). But unequal pairing and recombination of α or β with the homolog or sister chromatid can generate “loss-of-β” recombinants (Figure 5), which are easily identifiable among the progeny of crosses 2-4 by their nonparental pale aleurone phenotype. Rare kernels with the pale round nonparental phenotype were isolated as single-kernel events and putatively carry unequal recombination loss-of-β events. These candidate loss-of-β recombinants were confirmed using α- (primers XX390/QZ1265; Table 1) and β- (β-specific primers in Table 1) specific PCR primers to test for the presence of the 5.4-kb insertion and the absence of an intact β-component. Candidate recombinants were also confirmed by the segregation of pale kernels on selfed ears derived from the candidate pale kernels isolated in crosses 2–4.
Identification of recombination template and physical mapping of recombination breakpoints:
Homozygous recombinants were identified from among selfed progeny of confirmed recombination events isolated from crosses 2A–4A. Eight pale kernels were germinated for each confirmed event in 96-well flats and PCR-ready DNA was isolated as described (Dietrich et al. 2002). Seedlings homozygous for the recombinant allele were identified by genotyping these DNAs for the presence of the a1 allele provided by the male pollen parent of crosses 2A–4A. Seedlings that lacked this allele were selected for further analysis.
To determine the recombination template (i.e., homolog vs. sister chromatid) used to generate each recombinant from cross 2A, PCR-based markers within Sh2 were designed at single-nucleotide polymorphisms (SNPs) between the A1-b Sh2 and a1∷rdt Sh2 haplotypes. These polymorphisms were identified by temperature gradient capillary electrophoresis (TGCE) as described in Hsia et al. (2005) of Sh2-specific PCR products derived from A1-b Sh2 and a1∷rdt Sh2 genomic DNAs. Polymorphic PCR products were sequenced and PCR-based allele-specific markers designed at the polymorphic sites (Table 1).
To physically map unequal recombination breakpoints associated with a randomly chosen subset of recombinants isolated from crosses 2A–4A, primer sequences specific to the genomic subintervals α-Yz1A and β-Yz1B in the A1-b Sh2 haplotype (Figure 3A) and to a1∷rdt sh2 (GenBank accession no. AF072704) were designed at polymorphisms that exist among the sequences (Table 1).
For double-component recombinants generated by recombination between the β-Yz1B duplication and a1∷rdt-Yz1, all seedlings will be PCR positive for the rdt transposon insertion in exon 4 of a1. For those confirmed recombinant families for which the pollen parent in cross 2A was a1∷rdt sh2 and all eight seedlings were positive for the rdt insertion, 24 additional pale round kernels were germinated and DNA was isolated from resulting seedlings. Double-component recombinants were confirmed as those events for which all progeny tested positive for the rdt transposon insertion. The probability that a single-component recombinant homozygote is not identified in a population of 24 single-component recombinants is extremely low (i.e., 0.005%).
PCR conditions:
For interval-specific primers designed at SNPs, touchdown PCR was optimized with MgCl2 concentration and four different annealing temperatures (58°, 60°, 62°, or 64°). The PCR program consisted of 94° for 3 min; 10 cycles of 94° for 30 sec, 9° + annealing temperature for 45 sec (decrease by 0.8° per cycle), 72° for 1 min per every 1 kb of expected product; 25 cycles of 94° for 30 sec, annealing temperature for 45 sec, 72° for 1 min per every 1 kb of expected product; and a final extension at 72° for 10 min. For all other interval-specific primer pairs, the annealing temperature was optimized using a temperature gradient and the PCR program consisted of 94° for 3 min; 35 cycles of 94° for 30 sec, annealing temperature for 45 sec, 72° for 1 min per every 1 kb of expected product; and a final extension at 72° for 10 min.
Long-range PCR using TaKaRa Ex Taq polymerase (TaKaRa Bio, Japan) was conducted according to a web-published protocol (http://ppg.coafes.umn.edu/protocols.htm) from James Bradeen (University of Minnesota) on genomic DNAs prepared as described by Dellaporta et al. (1983). The modified PCR program consisted of 94° for 1 min; 14 cycles of 94° for 30 sec, 59° for 2 min, 72° for 15 min; 16 cycles of 94° for 30 sec, 59° for 2 min, 72° for 15 min (increase of 15 sec per cycle); and a final extension at 72° for 10 min. Resulting products were cloned using the TOPO TA cloning kit for sequencing (Invitrogen, San Diego), following manufacturer's instructions.
Identification and confirmation of intragenic recombination in a1 point mutant alleles:
Rates of intragenic recombination at a1 were measured using single-copy a1 alleles, each of which contained a single EMS-induced point mutation and conferred a colorless phenotype. In cross 5, the point mutant alleles were derived from two polymorphic A1 alleles (A1-Cornell and A1-Line C). In cross 6, the EMS-induced a1 alleles were derived from the same a1 allele (A1-Cornell) and the alleles differ only at the positions of the two EMS-induced lesions; this allele combination is referred to as “dimorphic” according to the nomenclature of Dooner (2002). The intervals assayed in the polymorphic and dimorphic allele combinations extend from positions 2438–2891 and 2435–2891, respectively, of the A1 sequence deposited in GenBank accession no. X05068:
Cross 5 (polymorphic): a1-3849-9 Sh2 et/a1-3845-5 Sh2 Et x a1-s Sh2 et/a1-s Sh2 et
Cross 6 (dimorphic): a1-3849-9 Sh2 et/a1-3849-5 Sh2 et x al-s Sh2 et/a1-s Sh2 et
As expected, most progeny from crosses 5 and 6 had parental colorless aleurone phenotypes. If intragenic recombination, however, occurred within the interval demarked by the two point mutations such that the recombinant restored a functional A1 allele (designated A1′), a colored kernel would result. For the cross involving the polymorphic allele combination (cross 5), crossover (CO) and noncrossover (NCO) intragenic recombinants are expected to have nonetched and etched aleurones, respectively. For the cross involving the dimorphic allele combination (cross 6), all CO and NCO recombinants are expected to have etched aleurones. Candidate intragenic recombinants were confirmed by detecting (via PCR) the a1-s allele contributed by the pollen parent and by the kernel phenotypes that segregated among the progeny of cross 7:
Cross 7: A1′Sh2/a1-s Sh2@
Statistical analyses:
χ2-Heterogeneity tests were used to compare rates of meiotic recombination. Distributions of recombination breakpoints across the a1-sh2 interval were compared with a combination of χ2-contingency and Freeman–Halton tests as previously described (Yao and Schnable 2005; Yandeau-Nelson et al. 2006).
RESULTS
A1-b is a duplication of the a1 gene:
On the basis of his genetic recombination experiments, Laughnan concluded that the A1-b locus consists of a tandem duplication of the a1 gene comprising two components: α and β (Laughnan 1949, 1952, 1955). Consistent with his conclusion, only two a1-hybridizing HindIII fragments (18 and 5.8 kb) are detected in A1-b genomic DNA (Figure 2, lane 4). In addition, similar gel blot analyses with EcoRI provide no evidence for the presence of more than two components within the A1-b duplication (data not shown). Using pulsed-field gel electrophoresis and DNA gel blot analyses of A1-b genomic DNA the physical distance between these two HindIII fragments was estimated to be ≤75 kb (data not shown). The 18-kb fragment, but not the 5.8-kb fragment, could be detected in several maize stocks in which the β-component had been replaced by another a1 allele (Figure 2, lanes 2, 3, and 5) via recombination (Neuffer 1965). These results demonstrate that the 18-kb and 5.8-kb fragments are derived from the α- and β-components, respectively. Both HindIII fragments were cloned and sequenced (materials and methods).
When separated from α via recombination, β confers a wild-type colored aleurone phenotype (Laughnan 1949, 1952). Consistent with this phenotype, β is structurally similar to wild-type A1 alleles. When separated from β via recombination, α confers a pale aleurone kernel phenotype (Laughnan 1952). Within the second intron of α is a complex 5.4-kb insertion flanked by 21-bp direct duplications. This insertion is composed of a 3.8-kb transposon-like element with 458-bp terminal inverted repeats (TIRs) and an adjacent 1.6-kb inverted partial duplication of a1 (Figure 3B). Chromosome rearrangements (e.g., inversions, deletions, and duplications) associated with transposons have been observed in Antirrhinum (reviewed in Coen et al. 1989; Martin and Lister 1989), Drosophila (Tsubota et al. 1989; Montgomery et al. 1991), and maize (McClintock 1942, 1950; Walker et al. 1995; Zhang and Peterson 1999; Slotkin et al. 2005). Alternatively spliced transcripts are produced by several mutant wx alleles that contain intronic retrotransposon insertions of sizes similar to the 5.4-kb insertion within α (Varagona et al. 1992; Marillonnet and Wessler 1997). Hence, the recessive pale aleurone phenotype associated with α may arise via a similar mechanism.
In a1-sh2 haplotypes containing a single-copy a1 gene, the yz1 gene is distal to a1 (Figure 1A). In the A1-b Sh2 haplotype, the yz1 gene could reside distal to α or distal to β or, if yz1 is also duplicated, copies of yz1 might be distal to both α and β. To determine if the yz1 gene is duplicated, long-range PCR was conducted using a primer specific to α (αIDP1R; Table 1) or a primer specific to β (β2149R; Table 1) paired with a yz1 primer (ZH1748; Table 1). Successful amplification and subsequent sequencing of ∼8-kb and ∼9.2-kb PCR fragments from α and β within the A1-b Sh2 haplotype established that two copies of the yz1 gene, termed Yz1A-1012M and Yz1B-1012M, reside ∼5.6 kb downstream of α and β, respectively (Figure 3A). Of this duplicated segment, the α- and β-components exhibit 96% sequence identity in the genic sequences flanking but not including the 5.4-kb insertion in α and at least 6.4 kb of the duplication, including yz1, exhibits 100% identity (Figure 3A).
Strategy to directly measure frequencies of interhomolog and interchromatid recombination at A1-b:
In homozygotes, unequal recombination can occur between both homologs and sister chromatids (i.e., interhomolog and interchromatid recombination; Figure 4A). Because the homolog and sister chromatid recombination templates cannot be unambiguously distinguished it is not possible to separately assay rates of interhomolog and interchromatid recombination in this genotype. In previous studies (Tovar and Lichtenstein 1992; Molinier et al. 2004), rates of interhomolog recombination were inferred by subtracting rates of recombination in hemizygotes, in which only sister chromatid recombination can occur (Figure 4C). To directly measure and compare interhomolog and interchromatid recombination rates, the general strategy used in this study (materials and methods; Figure 4) included the isolation of unequal recombinants from heterozygotes in which the tandem duplication was paired with a sequence-heterologous single-copy allele (Figure 4B). The structures of recombinant gametes generated via interhomolog recombination with the single-copy a1 homolog (Figure 4B) are molecularly distinguishable from those generated via interchromatid recombination events. This strategy therefore allows for direct and separate measurements of the rates and patterns of interhomolog and interchromatid recombination within a single genotype.
High rates of unequal recombination at A1-b are dependent upon the presence of a homolog:
Unequal pairing and recombination of the α-Yz1A or β-Yz1B duplications with the homolog or sister chromatid can generate recombinants that have lost the β-component of the A1-b duplication (Figure 5). Because the α- and β-components confer pale and colored aleurone phenotypes, respectively, recombinants from crosses 2A–4A (materials and methods) that arise in this manner can be identified by virtue of their nonparental pale kernel phenotype. Indeed, all of the putative pale recombinants failed to amplify with several β-specific primers (Table 1), confirming that the pale phenotype is associated with loss of β (or a large portion of it).
Rates of meiotic unequal recombination at A1-b were estimated in the presence of a homolog that contained either a sequence-heterologous single-copy a1 allele (a1∷rdt; cross 2A) or a sequence-identical duplicated haplotype (A1-b; cross 4A). Rates of unequal recombination at A1-b were not affected significantly (P-value >0.05) by the nature of the homolog (compare crosses 2A vs. 4A, Table 2). Recombinants that arose via unequal interchromatid recombination (similar to that depicted in Figure 5, class V) were isolated from cross 3A, in which A1-b was hemizygous over the entire a1-sh2 interval (Figure 1B). The rate of unequal recombination was ∼10- to 13-fold lower (P < 1.0 × 10−47) when a homolog was absent (i.e., when only the sister chromatid was available for recombination; cross 3A; Table 2) as compared to when a homolog was present (crosses 2A and 4A). In this study, rates of unequal recombination were assayed in a common genetic background (crosses 2A and 3A) whereas rates of recombination at A1-b reported in previous studies were generated from a combination of different A1-b haplotypes, a1 single-copy haplotypes, and genetic backgrounds. Even so, the fold-change relationships between rates derived from unequal recombination measured in this study are similar to previous estimates (Laughnan 1949, 1952). Hence, these observations provide additional support for the finding that at A1-b meiotic recombination events that generate recombinants in which the β-component is either lost or replaced by sequences from the homolog occur preferentially between homologs as opposed to sister chromatids.
Numbers of recombinants and rates of unequal recombination
Crossa . | Female genotypec . | No. isolated . | No. testedd . | No. confirmed . | No. correctede . | Population sizef . | Rate (× 10−4)g . |
|---|---|---|---|---|---|---|---|
| A. At A1-b | |||||||
| 2Ab | A1-b Sh2/a1∷rdt Sh2 | 432 | 342 | 223 | 282 | 459,400 | 6.1 ± 0.4 |
| 205 | 159 | 119 | 153 | 190,800 | 8.0 ± 0.6 | ||
| 3A | A1-b Sh2/ax-1 | 213 | 118 | 15 | 27 | 454,800 | 0.59 ± 0.14 |
| 4A | A1-b Sh2/A1-b Sh2 | 124 | 89 | 89 | 124 | 165,700 | 7.5 ± 0.7 |
| B. At A1-b(P415) | |||||||
| 2B | A1-b(P415) Sh2/a1∷rdt Sh2 | 450 | 343 | 212 | 278 | 418,700 | 6.6 ± 0.4 |
| 3B | A1-b(P415) Sh2/ax-1 | 466 | 329 | 194 | 275 | 338,900 | 8.1 ± 0.5 |
| 4B | A1-b(P415) Sh2/A1-b(P415) Sh2 | 24 | 21 | 13 | 15 | 39,100 | 3.8 ± 1.0 |
Crossa . | Female genotypec . | No. isolated . | No. testedd . | No. confirmed . | No. correctede . | Population sizef . | Rate (× 10−4)g . |
|---|---|---|---|---|---|---|---|
| A. At A1-b | |||||||
| 2Ab | A1-b Sh2/a1∷rdt Sh2 | 432 | 342 | 223 | 282 | 459,400 | 6.1 ± 0.4 |
| 205 | 159 | 119 | 153 | 190,800 | 8.0 ± 0.6 | ||
| 3A | A1-b Sh2/ax-1 | 213 | 118 | 15 | 27 | 454,800 | 0.59 ± 0.14 |
| 4A | A1-b Sh2/A1-b Sh2 | 124 | 89 | 89 | 124 | 165,700 | 7.5 ± 0.7 |
| B. At A1-b(P415) | |||||||
| 2B | A1-b(P415) Sh2/a1∷rdt Sh2 | 450 | 343 | 212 | 278 | 418,700 | 6.6 ± 0.4 |
| 3B | A1-b(P415) Sh2/ax-1 | 466 | 329 | 194 | 275 | 338,900 | 8.1 ± 0.5 |
| 4B | A1-b(P415) Sh2/A1-b(P415) Sh2 | 24 | 21 | 13 | 15 | 39,100 | 3.8 ± 1.0 |
Cross from which pale round recombinants were isolated.
Crosses 2A–4A were conducted in two separate isolation plots. For cross 2A, unlike in crosses 3A and 4A, the rates of unequal recombination differed significantly between plots and both rates are therefore presented.
The genotype from which pale round recombinants were isolated.
Putative pale recombinants were tested by genetic crosses and PCR analyses with the primers in Table 1.
No. corrected = no. isolated × (no. confirmed/no. tested).
Population sizes are based on the number of gametes containing the A1-b Sh2 chromosome.
Standard errors were calculated using the formula (pq/n)1/2.
Numbers of recombinants and rates of unequal recombination
Crossa . | Female genotypec . | No. isolated . | No. testedd . | No. confirmed . | No. correctede . | Population sizef . | Rate (× 10−4)g . |
|---|---|---|---|---|---|---|---|
| A. At A1-b | |||||||
| 2Ab | A1-b Sh2/a1∷rdt Sh2 | 432 | 342 | 223 | 282 | 459,400 | 6.1 ± 0.4 |
| 205 | 159 | 119 | 153 | 190,800 | 8.0 ± 0.6 | ||
| 3A | A1-b Sh2/ax-1 | 213 | 118 | 15 | 27 | 454,800 | 0.59 ± 0.14 |
| 4A | A1-b Sh2/A1-b Sh2 | 124 | 89 | 89 | 124 | 165,700 | 7.5 ± 0.7 |
| B. At A1-b(P415) | |||||||
| 2B | A1-b(P415) Sh2/a1∷rdt Sh2 | 450 | 343 | 212 | 278 | 418,700 | 6.6 ± 0.4 |
| 3B | A1-b(P415) Sh2/ax-1 | 466 | 329 | 194 | 275 | 338,900 | 8.1 ± 0.5 |
| 4B | A1-b(P415) Sh2/A1-b(P415) Sh2 | 24 | 21 | 13 | 15 | 39,100 | 3.8 ± 1.0 |
Crossa . | Female genotypec . | No. isolated . | No. testedd . | No. confirmed . | No. correctede . | Population sizef . | Rate (× 10−4)g . |
|---|---|---|---|---|---|---|---|
| A. At A1-b | |||||||
| 2Ab | A1-b Sh2/a1∷rdt Sh2 | 432 | 342 | 223 | 282 | 459,400 | 6.1 ± 0.4 |
| 205 | 159 | 119 | 153 | 190,800 | 8.0 ± 0.6 | ||
| 3A | A1-b Sh2/ax-1 | 213 | 118 | 15 | 27 | 454,800 | 0.59 ± 0.14 |
| 4A | A1-b Sh2/A1-b Sh2 | 124 | 89 | 89 | 124 | 165,700 | 7.5 ± 0.7 |
| B. At A1-b(P415) | |||||||
| 2B | A1-b(P415) Sh2/a1∷rdt Sh2 | 450 | 343 | 212 | 278 | 418,700 | 6.6 ± 0.4 |
| 3B | A1-b(P415) Sh2/ax-1 | 466 | 329 | 194 | 275 | 338,900 | 8.1 ± 0.5 |
| 4B | A1-b(P415) Sh2/A1-b(P415) Sh2 | 24 | 21 | 13 | 15 | 39,100 | 3.8 ± 1.0 |
Cross from which pale round recombinants were isolated.
Crosses 2A–4A were conducted in two separate isolation plots. For cross 2A, unlike in crosses 3A and 4A, the rates of unequal recombination differed significantly between plots and both rates are therefore presented.
The genotype from which pale round recombinants were isolated.
Putative pale recombinants were tested by genetic crosses and PCR analyses with the primers in Table 1.
No. corrected = no. isolated × (no. confirmed/no. tested).
Population sizes are based on the number of gametes containing the A1-b Sh2 chromosome.
Standard errors were calculated using the formula (pq/n)1/2.
Homolog is preferred over sister chromatid as recombination template:
To assess the rates at which interhomolog and interchromatid recombination occur when both templates are present, the recombination templates from which the recombinants in cross 2A were derived were determined. This was accomplished using PCR-based markers specific to the Sh2 allele in either the A1-b (i.e., sister chromatid repair; Figure 5, class V) or a1∷rdt (i.e., homolog repair; Figure 5, classes III and IV) haplotypes (materials and methods). From cross 2A, 281 randomly selected recombinants were genotyped with these markers.
Only 6% of these recombinants (N = 17) used the sister chromatid as the unequal recombination template (Figure 6A), demonstrating that the homolog is the preferred recombination template for the generation of pale recombinants. This rate (4.0 × 10−5) does not differ significantly (P = 0.15) from the rate of interchromatid recombination when only the sister chromatid is available (5.9 × 10−5; Table 2A) in cross 3A (similar to that depicted in Figure 5, class V). This comparison demonstrates that interhomolog recombination does not occur at the expense of interchromatid recombination.
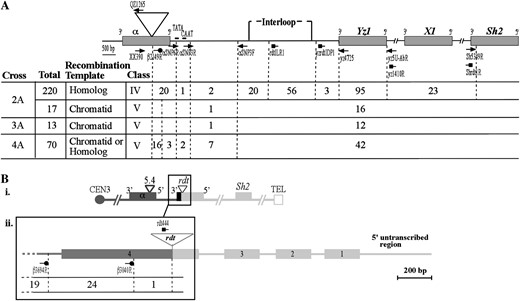
Physical mapping of recombination breakpoints. (A) Locations of unequal recombination breakpoints associated with single-component recombinants (Figure 5, classes IV and V). The schematic diagram represents a single-component recombinant haplotype in which boxes and lines represent genes and intergenic regions, respectively. The triangle indicates the position of the 5.4-kb insertion in α (not drawn to scale). The portion of the interval from the site of the 5.4-kb insertion through yz1, however, is drawn to scale. Thick solid horizontal bars designate the CAAT and TATA boxes (Tuerck and Fromm 1994; Pooma et al. 2002) of a1. The numbers of recombination breakpoints that resolved in each interval from each cross are indicated. Because the α-Yz1A and β-Yz1B intervals are sequence identical from the interloop region through at least Yz1 (Figure 3A), recombinants with the sister chromatid from crosses 2A–4A or with the homolog from cross 4A with recombination breakpoints that resolved distal to the position of the αSNP3F primer cannot be mapped to higher resolution. Crosses 2A–4A were conducted in two isolation plots. Although the rates of pale round recombinants recovered in cross 2A from the two isolation plots differed (see footnote b in Table 2A), χ2-contingency and Freeman–Halton tests failed to detect a significant difference between the distributions of recombination breakpoints from the two plots. Therefore, breakpoint distribution data from cross 2A were combined between the two plots. (B) Locations of unequal recombination breakpoints associated with double-component recombinants (Figure 5, class III) from cross 2A. (i) The structure of the double-component recombinant generated by pairing of the a1∷rdt homolog with β-Yz1B. The boxes and lines represent genes and intergenic regions, respectively. (ii) Locations of unequal recombination breakpoints proximal to the rdt insertion in exon 4. The schematic diagram represents the recombinant a1∷rdt component for which boxes and lines represent exons and introns, respectively. (A and B) Triangle, circle, and square arrows indicate positions of α-, β-, and a1∷rdt-specific primers used for PCR amplification, respectively.
Choice of pairing partner (α vs. β) with homolog is not random:
For recombinants from cross 2A that recombined with the a1∷rdt-containing homolog, the a1∷rdt-Yz1 interval could have paired with either the α-Yz1A or the β-Yz1B duplication. Pairing of and subsequent recombination between a1∷rdt-Yz1 and the α-Yz1A duplication produces “single-component” recombinants (Figure 5, class IV) that have lost β and retained a single recombinant α-component. Conversely, pairing of a1∷rdt-Yz1 with the β-Yz1B duplication generates “double-component” recombinants (Figure 5, class III) where α is retained and a1∷rdt (or a portion of it) replaced the β-component (or a portion of it). Single- and double-component recombinants generated by pairing of a1∷rdt-Yz1 with α-Yz1A or the β-Yz1B duplication were identified by genotyping for the absence or presence of the a1∷rdt allele, respectively.
Of the 94% of recombinants (N = 264) from cross 2A that used the homolog as recombination template, 83% (N = 220; Figure 6A) were generated by the pairing of a1∷rdt-Yz1 with α-Yz1A (Figure 5, class IV). Only 17% (N = 44) of recombinants occurred via pairing of the homolog with β-Yz1B (Figure 5, class III; Figure 6B). This demonstrates that the pairing configuration of the homolog with the duplicate components is not random but, instead, the a1∷rdt allele present on the homolog preferentially pairs with and recombines with the α-Yz1A duplication (Figure 5, class IV) as opposed to the β-Yz1B duplication (Figure 5, class III). For those recombinants generated by pairing of a1∷rdt-Yz1 with β-Yz1B to form double-component structures (Figure 5, class III), recombination breakpoints were mapped to higher resolution using component-specific PCR markers (Table 1; Figure 6B). Approximately half of the breakpoints in these double-component recombinants resolved within exon 4 of a1 and the remainder downstream of the a1 coding region (Figure 6B); this latter group was not mapped to higher resolution.
Molecular mapping of recombination events:
In Figure 5 only recombination breakpoints that resolve within a1 are illustrated. Recombination events could, however, potentially resolve anywhere within the region between the 5.4-kb insertion in α and the β-component (Figure 3A). To compare the distributions of recombination breakpoints across the recombinant α-Sh2 haplotype generated via recombination with different templates, recombination breakpoints from class IV and V recombinants (Figure 5) were mapped to higher resolution. Using PCR-based markers designed at sequence polymorphisms that exist among α, β, and a1∷rdt sequences (Table 1) across the a1-yz1 interval, breakpoints were mapped for class IV recombinants (Figure 5) with the homolog (cross 2A) or class V recombinants (Figure 5) with the sister chromatid (crosses 2A and 3A). In recombinants derived from A1-b homozygotes (cross 4A; similar to that depicted in Figure 5, class V), breakpoints were mapped but the recombination template associated with each recombinant could not be determined. Because both the rates (P-value = 0.15) and distribution of unequal recombination breakpoints (P-value = 0.811) with the sister chromatid did not differ between crosses 2A and 3A, in all subsequent analyses sister chromatid data from these two crosses were combined.
As has been observed in single-copy a1 haplotypes (Yao et al. 2002), the majority of recombination breakpoints associated with class IV and V recombinants (Figure 5) from crosses 2A–4A (Figure 6A) resolve within the a1-yz1 interval and cluster within previously identified recombination hot spots (i.e., a1, the interloop region, and yz1). Although the majority of breakpoints mapped in this study resolve within these hot spots, no breakpoints mapped to a1 (viz., the region demarked by the 5.4-kb insertion site and the CAAT box; Figure 6A) in interchromatid recombinants (crosses 2A and 3A; Figure 5, class V; Figure 6A), while ∼10% of recombinants map to this region in interhomolog recombinants with the a1∷rdt homolog (cross 2A; Figure 5, class IV; Figure 6A). Consistent with this difference, the distributions of breakpoints across the recombinant α-Sh2 haplotype generated either by interhomolog or by interchromatid recombination (cross 2A) differ significantly (P = 0.039).
Because very few recombinants use the sister chromatid as repair template (cross 2A; Figure 5, class V; Table 2A; Figure 6A), it is probable that most of the recombinants isolated from A1-b homozygotes (cross 4A) also resulted from interhomolog recombination. Consistent with this hypothesis, the distribution of breakpoints in cross 4A differs significantly from that of interchromatid recombinants (Figure 5, class V) isolated in crosses 2A and 3A (P-value = 0.013). The breakpoint distribution in the A1-b homozygote, however, also differs significantly from recombinants with the a1∷rdt homolog (Figure 5, class IV) in cross 2A (P-value = 9.1 × 10−8). In fact, 30% of unequal recombination breakpoints resolve within a1 in the A1-b homozygote (Figure 6A). These differences in breakpoint distributions suggest either that interchromatid recombination (Figure 5, class V) is more frequent in the A1-b homozygote or that recombinants resolve differently depending on pairing partner configuration (i.e., α- and a1∷rdt pairing in cross 2A vs. α- and β-pairing in cross 4A).
Rates of unequal interchromatid recombination are much higher in A1-b(P415) Sh2 than in A1-b Sh2:
Crosses were also conducted that were identical to crosses 1A–4A except that A1-b was replaced with A1-b(P415) Sh2 (crosses 1B–4B). Via genetic analyses, Laughnan demonstrated that like A1-b, A1-b(P415) also consists of α and β, but in the opposite order as compared to A1-b (i.e., centromere-β-α-Sh2 vs. centromere-α-β-Sh2; Laughnan 1955). Unlike the situation at A1-b, at A1-b(P415) the rate of unequal recombination (Table 2B) in the absence of a homolog (cross 3B) was actually higher than rates of recombination in the presence of a homolog (crosses 2B and 4B; P-value <0.02). While the rates of recombination in the presence of the a1∷rdt homolog did not significantly differ between A1-b and A1-b(P415) (Table 2; P-value >0.06), the rate of unequal interchromatid recombination (cross 3B) at A1-b(P415) was ∼13-fold higher than at A1-b (cross 3A; Table 2A; P-value = 9.0 × 10−65), demonstrating that unequal interchromatid recombination occurred more frequently in the A1-b(P415) genotype. Surprisingly, rates of unequal recombination also significantly differed between A1-b and A1-b(P415) homozygotes (crosses 4A and 4B, Table 2, A and B; P-value = 0.013). We cannot rule out the possibility that this difference is due to sampling variation as a consequence of the small population size in cross 4B that involved A1-b(P415) Sh2 (Table 2B).
Sequence heterology reduces recombination at a1 and might account for the increased rate of unequal interchromatid recombination at A1-b(P415):
The increased rate of unequal interchromatid recombination at A1-b(P415) (Table 2B) has been attributed to the opposite component order within A1-b(P415) as compared to A1-b (i.e., centromere-β-α-Sh2 vs. centromere-α-β-Sh2; Laughnan 1955). Partial sequencing (materials and methods) of A1-b(P415), however, reveals that the α- and β-components in A1-b(P415) differ not only in order as compared to A1-b, but also in their degree of sequence identity. In A1-b the α- and β-components share only 96.0% identity, while in A1-b(P415), α and β exhibit 99.9% identity, differing by only one 1-bp insertion (other than the complex insertion present in α) across the 2.5-kb sequenced region. Indeed, because the α- and β-components of A1-b(P415) are nearly identical, it was not possible to physically map the unequal recombination breakpoints associated with the pale recombinants isolated from this haplotype.
Although the order of components within a duplication might affect rates of unequal recombination, it is also possible that the higher amount of sequence identity between components in A1-b(P415) is stimulating unequal recombination rates between α and β when only the sister chromatid is present. Although this hypothesis is difficult to test between duplicated a1 alleles due to the technical challenges associated with gene replacement technology in plants that prevent the introduction of different levels of sequence identity between the components, the effect of sequence identity on rates of equal recombination can be easily assayed between single-copy a1 alleles.
To do so, the rate of intragenic recombination between polymorphic single-copy a1 alleles (cross 5) that exhibit 97% identity to each other was compared to the rate between dimorphic a1 alleles (cross 6) that differ at only two nucleotide positions (materials and methods). The a1 alleles each contained a single EMS-induced point mutation that rendered a1 nonfunctional and generated colorless aleurones. Intragenic recombination within the interval demarked by the lesions from the paired alleles can generate a recombinant allele via either a CO or a NCO that has “lost” the point mutations and restored a functional A1′ allele.
The rate of equal recombination between dimorphic a1 alleles (i.e., ∼100% identity; cross 6) was approximately sevenfold higher than the rate between polymorphic a1 alleles (cross 5) exhibiting ∼97% sequence identity (Table 3). Because the recombining interval was approximately the same size and in approximately the same location in both allele combinations, the observed increase in the rate of recombination between dimorphic alleles can be attributed to the higher level of sequence identity across that interval.
Rates of recombination between polymorphic and dimorphic EMS-induced a1 alleles
F1 heterozygote class . | . | . | No. isolated . | No. confirmeda . | Population size . | Recombination rate (cM)c . | ||
|---|---|---|---|---|---|---|---|---|
| Cross . | F1 allele combination . | Cl etb . | Cl Etb . | Cl et . | Cl Et . | |||
| Polymorphic | 5 | a1-3849-9 Sh2 et/a1-3845-5 Sh2 Et | 5 | 441 | 2 | 1d | 222,537 | 0.0013 ± 0.00008 |
| Dimorphic | 6 | a1-3849-9 Sh2 et/a1-3849-5 Sh2 et | 20 | 208 | 17 | 0 | 180,483 | 0.0094 ± 0.0002 |
F1 heterozygote class . | . | . | No. isolated . | No. confirmeda . | Population size . | Recombination rate (cM)c . | ||
|---|---|---|---|---|---|---|---|---|
| Cross . | F1 allele combination . | Cl etb . | Cl Etb . | Cl et . | Cl Et . | |||
| Polymorphic | 5 | a1-3849-9 Sh2 et/a1-3845-5 Sh2 Et | 5 | 441 | 2 | 1d | 222,537 | 0.0013 ± 0.00008 |
| Dimorphic | 6 | a1-3849-9 Sh2 et/a1-3849-5 Sh2 et | 20 | 208 | 17 | 0 | 180,483 | 0.0094 ± 0.0002 |
Intragenic recombinants were confirmed by a combination of genetic crosses and molecular analysis (e.g., genotyping for the a1 pollen parent).
Cl et, colored etched kernels; Cl Et, colored nonetched kernels.
Calculated as (no. confirmed Cl et + Cl Et)/population size × 100. Standard errors were calculated as described in Table 2, footnote g.
Two of the isolated Cl et candidates were PCR-positive for the pollen parent a1 allele. One was tested but not confirmed by genetic cross and the other died at the seedling stage. For the most conservative estimate of intragenic recombination, it is assumed that the dead seedling would have been confirmed and, therefore, there is one Cl et recombinant.
Rates of recombination between polymorphic and dimorphic EMS-induced a1 alleles
F1 heterozygote class . | . | . | No. isolated . | No. confirmeda . | Population size . | Recombination rate (cM)c . | ||
|---|---|---|---|---|---|---|---|---|
| Cross . | F1 allele combination . | Cl etb . | Cl Etb . | Cl et . | Cl Et . | |||
| Polymorphic | 5 | a1-3849-9 Sh2 et/a1-3845-5 Sh2 Et | 5 | 441 | 2 | 1d | 222,537 | 0.0013 ± 0.00008 |
| Dimorphic | 6 | a1-3849-9 Sh2 et/a1-3849-5 Sh2 et | 20 | 208 | 17 | 0 | 180,483 | 0.0094 ± 0.0002 |
F1 heterozygote class . | . | . | No. isolated . | No. confirmeda . | Population size . | Recombination rate (cM)c . | ||
|---|---|---|---|---|---|---|---|---|
| Cross . | F1 allele combination . | Cl etb . | Cl Etb . | Cl et . | Cl Et . | |||
| Polymorphic | 5 | a1-3849-9 Sh2 et/a1-3845-5 Sh2 Et | 5 | 441 | 2 | 1d | 222,537 | 0.0013 ± 0.00008 |
| Dimorphic | 6 | a1-3849-9 Sh2 et/a1-3849-5 Sh2 et | 20 | 208 | 17 | 0 | 180,483 | 0.0094 ± 0.0002 |
Intragenic recombinants were confirmed by a combination of genetic crosses and molecular analysis (e.g., genotyping for the a1 pollen parent).
Cl et, colored etched kernels; Cl Et, colored nonetched kernels.
Calculated as (no. confirmed Cl et + Cl Et)/population size × 100. Standard errors were calculated as described in Table 2, footnote g.
Two of the isolated Cl et candidates were PCR-positive for the pollen parent a1 allele. One was tested but not confirmed by genetic cross and the other died at the seedling stage. For the most conservative estimate of intragenic recombination, it is assumed that the dead seedling would have been confirmed and, therefore, there is one Cl et recombinant.
DISCUSSION
In A1-b, unequal interhomolog recombination occurs at higher rates than interchromatid recombination:
In meiotic recombination between equally paired genes, the homolog is the preferred recombination template (Petes and Pukkila 1995; reviewed in Kleckner 1996; Roeder 1997). Meiotic unequal interhomolog recombination occurred 10-fold more frequently than interchromatid recombination in a synthetic duplication of yeast (Jackson and Fink 1985). By comparing rates of meiotic unequal recombination in plants that contained and did not contain a homolog, Assaad and Signer (1992) and Tovar and Lichtenstein (1992) have generated indirect evidence that the homolog is the preferred template for meiotic unequal recombination in plants. A similar reasoning can be applied to analyses of the tandem duplication, A1-b, where rates of unequal recombination in the presence of the a1∷rdt homolog were 10- to 13-fold higher than when only the sister chromatid was present. But this study's molecular characterizations of recombination events also provide direct evidence that interhomolog recombination is preferred over interchromatid recombination (i.e., 94% of recombinants were interhomolog recombinants with the a1∷rdt homolog; Figure 5, classes III and IV; Figure 6). Interestingly, the low rate of interchromatid recombination observed in A1-b/a1∷rdt heterozygotes is very similar to the rate of unequal recombination in A1-b hemizygotes. To our knowledge, this is the first molecular analysis of unequal interchromatid recombination both in the absence and in the presence of a homolog and demonstrates that at A1-b interhomolog recombination does not occur at the expense of interchromatid recombination.
In A1-b(P415), are rates of unequal interhomolog and interchromatid recombination similar?
The situation is quite different at A1-b(P415) where the rate of unequal recombination in the presence of the homolog (cross 2B) is actually lower than in its absence (cross 3B). Although rates of interhomolog and interchromatid recombination could not be directly and separately measured, this result suggests that in A1-b(P415) the presence of a homolog actually suppresses interchromatid recombination. In addition, interchromatid recombination in A1-b(P415) hemizygotes occurs at a rate ∼13-fold more frequently than interchromatid recombination at A1-b (Table 2). In combination, these results demonstrate that interchromatid recombination at A1-b(P415) is regulated differently than at A1-b.
Regulation of recombination at A1-b and A1-b(P415):
The difference in interchromatid recombination rates experienced by A1-b and A1-b(P415) was previously attributed to the opposite orders of the α- and β-components (Laughnan 1955) in these two haplotypes. This study, however, identified a second structural difference between the two haplotypes: the much higher sequence identity between the components of A1-b(P415) than those of A1-b (99.9% vs. 96.0%). Sequence similarity is, in general, correlated with higher rates of recombination (reviewed in Modrich and Lahue 1996; Paques and Haber 1999). For example, in the maize bz1 gene rates of recombination were approximately twofold higher between nearly identical bz1 alleles than between alleles exhibiting ∼1.5% divergence (Dooner 2002). Similarly, at a synthetic tandemly arrayed gene cluster in Arabidopsis, meiotic unequal interchromatid recombination events tended to resolve in regions of higher sequence identity between the duplicated sequences (Jelesko et al. 1999, 2004). In yeast, rates of meiotic recombination were significantly reduced by increased sequence divergence between inverted repeat sequences (Chen and Jinks-Robertson 1999). Similarly, in somatic cells in Arabidopsis, only 0.16% divergence between repeats reduced the rate of unequal recombination threefold (Opperman et al. 2004).
Although the effects of sequence divergence between A1-b components on meiotic unequal recombination were not tested directly, the rate of equal recombination between nearly identical alleles of single-copy a1 homologs was sevenfold higher than between alleles exhibiting ∼3% divergence (dimorphic vs. polymorphic; Table 3). This observation is consistent with the high rate of unequal recombination between the nearly identical α- and β-components in A1-b(P415) as compared to the lower rate seen between the 4% divergent components in A1-b. Because the alleles in the polymorphic combination (Table 3) are derived from different genetic backgrounds, we cannot rule out the possibility that genetic background may be affecting recombination.
Surprisingly, only in the absence of a homolog (crosses 3A and 3B; Table 2) were rates of unequal recombination significantly higher in A1-b(P415) than in A1-b. These results suggest that there is a hierarchy of factors affecting rates of unequal interhomolog and interchromatid recombination. First, during meiosis the homolog is the preferred template for unequal recombination within a tandem duplication and this choice of template is not dependent on the level of sequence polymorphism between homologs. In yeast, several proteins (e.g., DMC1, TID1, RED1, MEK1, HOP1, and RDH54) specifically promote meiotic equal recombination (Klein 1997; Schwacha and Kleckner 1997; Arbel et al. 1999; Thompson and Stahl 1999; Niu et al. 2005) and might likewise act to promote meiotic unequal interhomolog recombination. In fact, DMC1 plays a role in the preference for unequal interhomolog recombination during meiosis (Thompson and Stahl 1999). If, however, only the sister chromatid is available for repair (crosses 3A and 3B), rates of unequal recombination are correlated with sequence identity between the components of the duplication. This hypothesis is consistent with the finding that rates of unequal recombination were increased between the nearly identical components of the A1-b(P415) duplication in the absence of a homolog and, hence, between sister chromatids. We cannot, however, rule out the presence of polymorphic structures outside of the duplication that might differentially affect interchromatid recombination in the A1-b and A1-b(P415) genetic stocks.
Although the increased sequence identity between components of A1-b(P415) can adequately explain the increased rate of interchromatid recombination in A1-b(P415) hemizygotes as compared to A1-b hemizygotes, the observation that the rate of unequal recombination in A1-b(P415) hemizygotes exceeds the rate when A1-b(P415) is paired with the a1∷rdt homolog (Table 2) is puzzling. A possible explanation for this unexpected result is that another type of recombination event that yields nonviable products (Figure 5) competes with unequal interhomolog and interchromatid recombination in some haplotypes. Alternatively, polymorphic structures flanking the a1 locus in A1-b(P415) and the a1∷rdt Sh2 homolog might act together to suppress rates of unequal recombination in the presence of that homolog.
The choice of unequal pairing partner with the homolog is not random:
Unequal recombination at A1-b occurs preferentially between homologs and a single-copy a1 homolog can pair with either component within the A1-b duplication. In 83% of the recombinants from cross 2 the a1∷rdt homolog paired with the α-Yz1A duplication (Figure 5, class IV) rather than with the β-Yz1B duplication (Figure 5, class III), thereby demonstrating that pairing partners are not selected at random. This finding explains Laughnan's (1952) observation that one phenotypic class of unequal recombinants occurred much more frequently than another.
Since a higher level of sequence identity appears to contribute to the increased rate of unequal interchromatid recombination, it could be hypothesized that the level of sequence identity might similarly affect the choice of pairing partner (i.e., α vs. β) with the homolog. Our results, however, are inconsistent with this hypothesis. Although the a1∷rdt homolog exhibits a higher level of sequence identity with β (97.3%) than with α (95.2%, which does not include the 5.4-kb insertion within α), a1∷rdt pairs with and recombines with the α-Yz1A duplication more frequently than with the β-Yz1B duplication. This demonstrates that the choice of pairing partner is most likely not associated with the degree of sequence polymorphism between the a1 alleles. Instead, chromatin structure or cis-acting elements polymorphic between the components may influence pairing preference.
This observed preference for pairing partner has the potential to influence gene loss/gain and diversity within tandemly duplicated gene arrays. For example, in the A1-b duplication the preference for the homolog to pair with the α-Yz1A segment resulted in recombinant haplotypes that underwent gene loss (Figure 5, class IV). If, however, the alternative pairing configuration (in this case, a1∷rdt with the β-Yz1B duplication) was preferred, the majority of recombinants would contain duplications of a1 (Figure 5, class III), which could, via subsequent rounds of unequal recombination, expand into tandem arrays with increasing copies of a1.
Recombination breakpoint resolution sites are affected by recombination template:
Because rates of recombination are affected by both choice of recombination template (homolog vs. sister chromatid) and pairing partner (α vs. β) with the homolog, these factors might also affect distributions of recombination breakpoints associated with unequal recombination events. To our knowledge, this is the first study in which the distribution of recombination breakpoints associated with unequal interhomolog and interchromatid recombination can be compared.
The distribution of unequal recombination breakpoints (Figure 6A) associated with interhomolog recombination (cross 2A; Figure 5, class IV) differed significantly from that of interchromatid recombination (crosses 2A and 3A; Figure 5, class V; Figure 6A). Further, distributions of breakpoints associated with recombination between A1-b and the a1∷rdt homolog (cross 2A) differed significantly as compared to the distribution of breakpoints in the A1-b Sh2 homozygote (cross 4A; Figure 6A). Because most recombinants isolated from the A1-b Sh2/a1∷rdt Sh2 F1 occurred via interhomolog recombination (Figure 5, classes III and IV), it is reasonable to assume that the preferred recombination template in the A1-b homozygote is also the homolog. In each case, differences in breakpoint distributions might be attributed to polymorphic cis-acting elements that affect unequal recombination between A1-b Sh2 and a1∷rdt Sh2 homologs as compared to recombination between A1-b Sh2 sister chromatids. Indeed, cis-acting elements have been previously shown to affect recombination across the a1-sh2 interval (Yao and Schnable 2005). Alternatively, components of the recombination machinery specific to unequal interhomolog recombination (Thompson and Stahl 1999) might differently affect the resolution of recombination breakpoints in homologs as compared to sister chromatids. This is supported by the observation that recombination breakpoint distributions differ between A1-b sister chromatids (cross 3A) and A1-b homozygotes (cross 4A; Figure 6A).
Unequal recombination at A1-b occurs at rates similar to equal recombination at single-copy a1 alleles:
Previous molecular analyses of unequal recombination have been conducted using synthetic duplications (Jelesko et al. 1999, 2004; Molinier et al. 2004). This is the first in-depth molecular analysis of a naturally occurring tandem duplication. In contrast to synthetic duplications, patterns of recombination at the endogenous duplicate A1-b allele can be directly compared to patterns of recombination at that same genomic location in a haplotype containing a single-copy allele (i.e., a1).
Rates of unequal recombination between the a1∷rdt homolog and either A1-b or A1-b(P415) (Table 2; ∼10−4) did not differ significantly (P-value >0.16) from rates of interhomolog recombination (7.0 × 10−4) across a1-Yz1 intervals that contain single-copy a1 alleles (Yao et al. 2002). The rates of A1-b recombination reported here, however, represent lower limits. First, the assay employed in this study does not detect equal recombination events that resolve telomeric to the distal component in the A1-b and A1-b(P415) duplications. Second, measured rates of unequal recombination represent only events that resulted in a change in aleurone phenotype (i.e., recombinants in which β was either lost or replaced by the homolog). Therefore, rates of recombination at the tandem duplication of a1 very likely exceed rates previously determined in single-copy a1-sh2 haplotypes and suggest the possibility that rates of recombination might further increase with increasing numbers of components within a tandem duplication.
The use of a naturally occurring duplication also allowed us to determine that the distribution of recombination breakpoints in A1-b is similar to distributions observed in single-copy haplotypes; i.e., recombination hot spots are conserved. Therefore, at least some of the factors that determine meiotic recombination hot spots act in both equal and unequal recombination.
Evolutionary and breeding implications:
This study extends our knowledge of factors that affect unequal recombination between tandemly duplicated genes. Relatively high rates of unequal recombination coupled with definite pairing preferences (summarized in Table 4) can greatly affect and direct the evolution of tandemly arrayed duplicate gene families within the maize genome. Because approximately one-third of maize genes are members of tandemly arrayed duplicate gene families (Blanc and Wolfe 2004; Messing et al. 2004), the process of unequal recombination between duplicated sequences could have profound effects on genome evolution. The number of gene copies in a tandemly arrayed duplicate gene family is likely to vary among haplotypes as a consequence of unequal recombination. Even within inbreds, unequal recombination between components of a duplication can generate diversity. Indeed, diversity within long-term inbred lines in maize has been observed both molecularly (Gethi et al. 2002; Heckenberger et al. 2002) and phenotypically (Busch and Russell 1964; Fleming et al. 1964; Russell et al. 1963; Russell and Vega 1973). While such diversity is often attributed to residual heterozygosity, mutation, or pollen contamination, another possible mechanism is unequal recombination within the many tandemly arrayed duplicate gene families of maize. Such variation within the inbred line B73 (the inbred selected for genome sequencing) could potentially influence the universality of the maize genome sequence.
Summary of recombination at A1-b and A1-b(P415)
. | α and β . | Rate of recombination in the absence vs. presence of homolog . | . | Preferred pairing partner with homolog . | |
|---|---|---|---|---|---|
| Haplotype . | Order . | Sequence identity (%) . | Preferred template . | ||
| A1-ba | α-β-Sh2 | 96 | 10- to 13-fold lower | 94% paired with homolog | α |
| A1-b(P415)a | β-α-Sh2 | 99.9 | 1.2-fold higher | NDb | NDc |
. | α and β . | Rate of recombination in the absence vs. presence of homolog . | . | Preferred pairing partner with homolog . | |
|---|---|---|---|---|---|
| Haplotype . | Order . | Sequence identity (%) . | Preferred template . | ||
| A1-ba | α-β-Sh2 | 96 | 10- to 13-fold lower | 94% paired with homolog | α |
| A1-b(P415)a | β-α-Sh2 | 99.9 | 1.2-fold higher | NDb | NDc |
The rates of recombination in the presence of a homolog are similar between A1-b and A1-b(P415). In the absence of a homolog (i.e., in a hemizygote), however, the rate of recombination is ∼13-fold higher in A1-b(P415) as compared to A1-b.
The preferred template in the presence of a homolog could not be determined in A1-b(P415).
Because α and β are nearly identical, the pairing partner cannot be determined.
Summary of recombination at A1-b and A1-b(P415)
. | α and β . | Rate of recombination in the absence vs. presence of homolog . | . | Preferred pairing partner with homolog . | |
|---|---|---|---|---|---|
| Haplotype . | Order . | Sequence identity (%) . | Preferred template . | ||
| A1-ba | α-β-Sh2 | 96 | 10- to 13-fold lower | 94% paired with homolog | α |
| A1-b(P415)a | β-α-Sh2 | 99.9 | 1.2-fold higher | NDb | NDc |
. | α and β . | Rate of recombination in the absence vs. presence of homolog . | . | Preferred pairing partner with homolog . | |
|---|---|---|---|---|---|
| Haplotype . | Order . | Sequence identity (%) . | Preferred template . | ||
| A1-ba | α-β-Sh2 | 96 | 10- to 13-fold lower | 94% paired with homolog | α |
| A1-b(P415)a | β-α-Sh2 | 99.9 | 1.2-fold higher | NDb | NDc |
The rates of recombination in the presence of a homolog are similar between A1-b and A1-b(P415). In the absence of a homolog (i.e., in a hemizygote), however, the rate of recombination is ∼13-fold higher in A1-b(P415) as compared to A1-b.
The preferred template in the presence of a homolog could not be determined in A1-b(P415).
Because α and β are nearly identical, the pairing partner cannot be determined.
Footnotes
Present address: Pennsylvania State University, University Park, PA 16802.
Present address: Donald Danforth Plant Science Center, St. Louis, MO 63132.
Footnotes
Communicating editor: A. H. Paterson
Acknowledgement
We thank the late John R. Laughnan for encouraging this research direction, James Bradeen (University of Minnesota) for sharing his long-range PCR protocol and related advice, and undergraduate students Brian Brand, Timothy Dunham, Ryan Manser, John Tenhunfeld, and George Weber for technical assistance. This research was supported in part by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service, grant nos. 9701407 and 9901579 to P.S.S. and Basil J. Nikolau and 0101869, 0300940, and 0500962 to P.S.S.; additional support was provided by Hatch Act and State of Iowa funds.
References
Allers, T., and M. Lichten,
Arabidopsis Genome Initiative,
Arbel, A., D. Zenvirth and G. Simchen,
Assaad, F. F., and E. R. Signer,
Bhave, M. R., S. Lawrence, C. Barton and L. C. Hannah,
Blanc, G., and K. H. Wolfe,
Busch, R. H., and W. A. Russell,
Chen, W., and S. Jinks-Robertson,
Chin, D. B., R. Arroyo-Garcia, O. E. Ochoa, R. V. Kesseli, D. O. Lavelle et al.,
Civardi, L., Y. Xia, K. J. Edwards, P. S. Schnable and B. J. Nikolau,
Coen, E. S., T. P. Robbins, J. Almeida, A. Hudson and R. Carpenter,
da Costa e Silva, O., R. Lorbiecke, P. Garg, L. Muller, M. Wassmann et al.,
Das, O. P., E. Poliak, K. Ward and J. Messing,
Dellaporta, S. L., J. Wood and J. B. Hicks,
Dietrich, C. R., F. Cui, M. L. Packila, J. Li, D. A. Ashlock et al.,
Dooner, H. K.,
Dooner, H. K., and J. L. Kermicle,
Dooner, H. K., and J. L. Kermicle,
Dooner, H. K., and I. M. Martinez-Ferez,
Fleming, A. A., G. M. Kozelnicky and E. B. Browne,
Garcia-Fernandez, J.,
Gethi, J. G., J. A. Labate, K. R. Lamkey, M. E. Smith and S. Kresovich,
Gonzalez-Barrera, S., F. Cortes-Ledesma, R. E. Wellinger and A. Aguilera,
Heckenberger, M., M. Bohn, J. S. Ziegle, L. K. Joe, J. D. Hauser et al.,
Hsia, A. P., and P. S. Schnable,
Hsia, A. P., T. J. Wen, H. D. Chen, Z. Liu, M. D. Yandeau-Nelson et al.,
International Rice Genome Sequencing Project, I. R. G. S.,
Jackson, J. A., and G. R. Fink,
Jackson, J. A., and G. R. Fink,
Jelesko, J. G., R. Harper, M. Furuya and W. Gruissem,
Jelesko, J. G., K. Carter, W. Thompson, Y. Kinoshita and W. Gruissem,
Johnson, R. D., and M. Jasin,
Kadyk, L. C., and L. H. Hartwell,
Kleckner, N.,
Klein, H. L.,
Kuang, H., S. S. Woo, B. C. Meyers, E. Nevo and R. W. Michelmore,
Laughnan, J. R.,
Laughnan, J. R.,
Laughnan, J. R.,
Leister, D.,
Lowe, B., J. Mathern and S. Hake,
Marillonnet, S., and S. R. Wessler,
Martin, C., and C. Lister,
McClintock, B.,
McClintock, B.,
Messing, J., A. K. Bharti, W. M. Karlowski, H. Gundlach, H. R. Kim et al.,
Modrich, P., and R. Lahue,
Molinier, J., G. Ries, S. Bonhoeffer and B. Hohn,
Montgomery, E. A., S. M. Huang, C. H. Langley and B. H. Judd,
Neuffer, M. G.,
Niu, H., L. Wan, B. Baumgartner, D. Schaefer, J. Loidl et al.,
Opperman, R., E. Emmanuel and A. A. Levy,
Paques, F., and J. E. Haber,
Petes, T. D., and P. J. Pukkila,
Pooma, W., C. Gersos and E. Grotewold,
Ramakrishna, W., J. Emberton, M. Ogden, P. SanMiguel and J. L. Bennetzen,
Richter, T. E., T. J. Pryor, J. L. Bennetzen and S. H. Hulbert,
Robbins, T. P., E. L. Walker, J. L. Kermicle, M. Alleman and S. L. Dellaporta,
Roeder, G. S.,
Russell, W. A., and U. A. Vega,
Russell, W. A., G. F. Sprague and L. H. Penny,
Sambrook, J., E. F. Fritsch and T. Maniatis,
Schwacha, A., and N. Kleckner,
Schwarz-Sommer, Z., N. Shepherd, E. Tacke, A. Gierl, W. Rohde et al.,
Semple, C., and K. H. Wolfe,
Slotkin, R. K., M. Freeling and D. Lisch,
Smith, S. M., and S. H. Hulbert,
Stadler, L. J., and M. G. Nuffer,
Stadler, L. J., and H. Roman,
Stinard, P. S., and D. S. Robertson,
Sturtevant, A. H.,
Sudupak, M. A., J. L. Bennetzen and S. H. Hulbert,
Sun, H., D. Treco and J. W. Szostak,
Sun, Q., N. C. Collins, M. Ayliffe, S. M. Smith, J. Drake et al.,
Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein and F. W. Stahl,
Thompson, D. A., and F. W. Stahl,
Tovar, J., and C. Lichtenstein,
Tsubota, S. I., D. Rosenberg, H. Szostak, D. Rubin and P. Schedl,
Tuerck, J. A., and M. E. Fromm,
Varagona, M. J., M. Purugganan and S. R. Wessler,
Walker, E. L., T. P. Robbins, T. E. Bureau, J. Kermicle and S. L. Dellaporta,
Webb, C. A., T. E. Richter, N. C. Collins, M. Nicolas, H. N. Trick et al.,
Williams, S. M., and L. G. Robbins,
Xu, X., A. P. Hsia, L. Zhang, B. J. Nikolau and P. S. Schnable,
Yandeau-Nelson, M. D., B. J. Nikolau and P. S. Schnable,
Yao, H., and P. S. Schnable,
Yao, H., Q. Zhou, J. Li, H. Smith, M. Yandeau et al.,
Yao, H., L. Guo, Y. Fu, L. A. Borsuk, T. J. Wen et al.,
Zhang, F., and T. Peterson,
Zhang, J.,
Zhang, J., and T. Peterson,
Zhang, L., and B. S. Gaut,



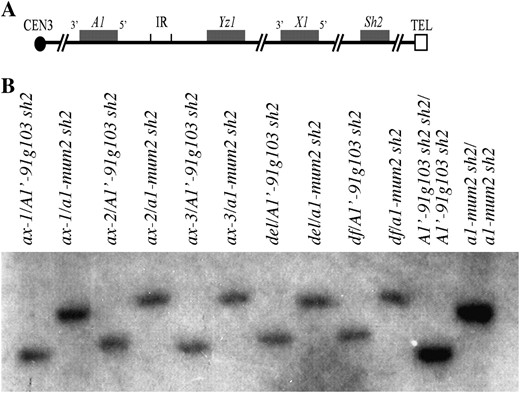
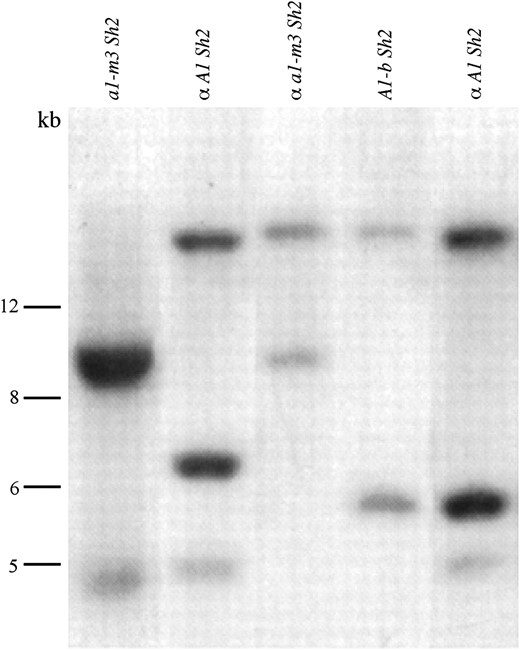
![Structure of the A1-b Sh2 haplotype. (A) The A1-b Sh2 haplotype. The sequence identities and sizes of duplicated segments are shown. Black lines designate regions with 100% sequence identity. Boxes and lines designate genes and intergenic regions, respectively. It is not known whether the x1 gene [which is ∼35 kb distal to yz1 in nonduplicated haplotypes (Yao et al. 2002)] is included in the duplication associated with the A1-b haplotype. Not drawn to scale. IR, interloop region (Yao et al. 2002). (B) Structure of A1-b α. A 5.4-kb insertion containing a 3.8-kb transposon-like element that contains a nested 453-bp Ins2 element with 93% identity to Ins2 in bz-R (GenBank accession no. X07938) is located within intron 2. The 3.8-kb element is flanked by 458-bp terminal inverted repeats (TIRs; long thick arrows), for which the first 13 bp of the TIRs are identical to the 13-bp TIRs of Ins2 (short arrows). The distal 458-bp TIR is actually a part of an intact 636-bp transposable element (triangle) with 93% identity to an uncharacterized transposon in intron 3 of the maize fatty aldehyde dehydrogenase1 gene (GenBank accession no. AY374447). Directly adjacent to the 3.8-kb element is a 1.6-kb inverted duplication of a1 sequence (shown in boldface type). Flanking the 5.4-kb insertion are 21-bp flanking direct duplications (solid bars). A Cin4 retrotransposon insertion is located in exon 4 at the same position as within other type II A1 alleles (Schwarz-Sommer et al. 1987) but is not present in the A1-b β-component. Boxes and lines represent exons and introns, respectively. Triangles represent insertions and arrows identify TIRs.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/genetics/173/4/10.1534_genetics.105.052712/3/m_2211fig3.jpeg?Expires=1716845365&Signature=MmhQqmqL3MZfony8jvMDOt13OHdpJ97rum2HZ-qP0Jy7yVTluppKlPwcHokUfyn2sq4Dx4HVp0Nss4kZJBYE4qD~5EiTgzz-uSz2o6fDj8OKAi85jdE~2~jgpiMMP038H~WRGrS0QjdMCBSseaku7-4k-81npMT56qWqV~7rt8c5DvIgc2IpBkION4wEHVnceEFQYgtDSTOrgUVvTlBxKvhuGSjFD8J7lxVS1vRlQ1NpqwtfzelYAGRnXsEMIPgFe4jyjodVbyhlDDPy63GN0k~Ik7nlihHQNWlp5IOtymJUlD5weUSUWTUvrljD9jVIv5NbbpN5nOyc3ef8qwbDxA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)