-
PDF
- Split View
-
Views
-
Cite
Cite
Craig J Ceol, Frank Stegmeier, Melissa M Harrison, H Robert Horvitz, Identification and Classification of Genes That Act Antagonistically to let-60 Ras Signaling in Caenorhabditis elegans Vulval Development, Genetics, Volume 173, Issue 2, 1 June 2006, Pages 709–726, https://doi.org/10.1534/genetics.106.056465
Close - Share Icon Share
Abstract
The synthetic multivulva (synMuv) genes negatively regulate Ras-mediated vulval induction in the nematode Caenorhabditis elegans. The synMuv genes define three classes, A, B, and C, such that double mutants carrying mutations in genes of any two classes are multivulva. The class B synMuv genes include lin-35, a homolog of the retinoblastoma (Rb) tumor suppressor gene, as well as homologs of genes that function with Rb in transcriptional regulation. We screened for additional synMuv mutations using a strategy different from that of previous synMuv genetic screens. Some of the mutations we recovered affect new synMuv genes. We present criteria for assigning synMuv mutations into different genetic classes. We also describe the molecular characterization of the class B synMuv gene lin-65.
A fundamental issue in developmental biology is how cells that are initially equivalent in developmental potential ultimately adopt different fates. Genetic studies have indicated that cells within a developmental equivalence group often adopt different fates in response to the combined action of multiple and sometimes competing signals (reviewed by Freeman and Gurdon 2002). For example, the initial step of R8 photoreceptor specification in ommatidial development in Drosophila melanogaster uses both positive and negative signals to properly select presumptive R8 photoreceptors from a field of developmentally equivalent cells in the eye imaginal disc (reviewed by Frankfort and Mardon 2002). An overlay of such signals can make a response in binary cell-fate decisions more precise or can increase the number of fates available to a particular cell.
Vulval development in the nematode Caenorhabditis elegans involves a set of ectodermal Pn.p cells that initially have similar developmental potentials but ultimately adopt different fates (Kimble 1981; Sternberg and Horvitz 1986). The specification of Pn.p cells that eventually make vulval tissue occurs in two steps, each of which involves the selection of a subset of Pn.p cells from a larger Pn.p field (Sulston and Horvitz 1977). First, in the L1 larval stage shortly after the 12 Pn.p cells are generated, the P1.p and P2.p anterior and P(9–11).p posterior cells fuse with the syncytial hypodermis. Each of the six remaining unfused midbody cells P(3–8).p has the capacity to adopt a vulval cell fate (Sternberg and Horvitz 1986). Second, three of these six cells, P(5–7).p, adopt vulval fates and undergo three rounds of division to generate seven (P5.p and P7.p) or eight (P6.p) descendants. P3.p, P4.p, and P8.p adopt nonvulval fates, typically dividing only once to generate two descendants that eventually fuse with the syncytial hypodermis. The decision to adopt vulval cell fates occurs during the L2 and early L3 larval stages and is followed by cell divisions and differentiation in the L3 and L4 larval stages, respectively (Kimble 1981; Sternberg and Horvitz 1986; Ferguson et al. 1987).
Many genes that control the specification of Pn.p fates have been identified. Some of these genes act in a spatially restricted fashion to select Pn.p cells for vulval development. The homeobox gene lin-39 is expressed in the midbody and regulates the sequential steps of fusion and vulval cell-fate specification of the Pn.p cells in this region (Clark et al. 1993; Wang et al. 1993; Maloof and Kenyon 1998). Strong loss-of-function lin-39 mutations result in ectopic P(3–8).p cell fusion during the L1 stage. In partial loss-of-function lin-39 mutants, unfused P(5–7).p cells are sometimes observed and often show vulval-to-non-vulval cell-fate transformations (Clark et al. 1993). lin-39 activity therefore promotes unfused cell fates in the L1 stage and vulval cell fates in the L2 and early L3 stages. Genes in the let-60 Ras signaling pathway also regulate the specification of Pn.p fates (Beitel et al. 1990; Han and Sternberg 1990). In addition to let-60 Ras, this pathway includes the receptor tyrosine kinase let-23, the SH2/SH3 adaptor sem-5, and the MAP kinase mpk-1, all of which are broadly conserved in Ras signaling systems (reviewed by Moghal and Sternberg 2003). The role of let-60 Ras signaling in the specification of vulval cell fates is well characterized. In wild-type animals, the let-60 Ras pathway is specifically activated in P(5–7).p in response to an EGF-like signal, encoded by lin-3, that is produced by the neighboring gonadal anchor cell (Hill and Sternberg 1992). Mutations that reduce let-60 Ras pathway activity prevent P(5–7).p vulval cell-fate specification, resulting in a vulvaless (Vul) phenotype. Mutations that abnormally activate this pathway cause P(3–8).p all to adopt vulval cell fates, resulting in a multivulva (Muv) phenotype (Beitel et al. 1990; Han and Sternberg 1990; Eisenmann and Kim 1997). Increases in let-60 Ras pathway activity may promote vulval cell fates in part by upregulating lin-39 expression (Maloof and Kenyon 1998).
The activities of lin-39 and genes in the let-60 Ras pathway are antagonized by the synthetic multivulva (synMuv) genes. The synMuv genes define three redundant classes, A, B, and C (Ferguson and Horvitz 1989; Thomas et al. 2003; Ceol and Horvitz 2004). Animals carrying mutations affecting any two classes of synMuv genes are Muv, but animals with a mutation in one synMuv gene or in multiple synMuv genes of a single class undergo wild-type vulval development. All three classes of genes promote the expression of nonvulval cell fates by P(3–8).p. At present it is unknown whether the synMuv mutations cause an increase of let-60 Ras pathway activity in these cells or cause these cells to be more sensitive to normal levels of let-60 Ras pathway activity. Roles for synMuv genes in regulating Pn.p fusion have also been described. Some class B genes, but no class A genes, antagonize lin-39-mediated cell fusion of at least one Pn.p cell, P3.p (Chen and Han 2001).
Many synMuv genes have been molecularly characterized. The class B synMuv protein LIN-35 is similar to the mammalian tumor suppressor pRb (Lu and Horvitz 1998). Other class B synMuv proteins include DPL-1 and EFL-1, which are similar to mammalian DP and E2F proteins and, by analogy to their mammalian counterparts, likely function to target LIN-35 retinoblastoma (Rb) to DNA (Ceol and Horvitz 2001). The class B synMuv protein HDA-1 is similar to class I histone deacetylases (Lu and Horvitz 1998) and may be targeted to specific genes by a DPL-1/EFL-1/LIN-35-containing protein complex.
As lin-35 Rb can act in the surrounding hypodermis to regulate P(3–8).p fates, the genes targeted by a DPL-1/EFL-1/LIN-35-containing complex may function non-cell autonomously to regulate the specification of vulval cell fates (Myers and Greenwald 2005). Other class B synMuv proteins also are components of this complex (M. M. Harrison and H. R. Horvitz, unpublished observations), and complexes purified from Drosophila extracts containing DP, E2F, and Rb homologs contain homologs of the synMuv proteins LIN-9, LIN-37, LIN-52, LIN-53 RbAp48, LIN-54, and LIN-61 (Korenjak et al. 2004; Lewis et al. 2004). These Drosophila complexes can repress transcription of DP/E2F target genes and can inhibit genomewide DNA replication in ovarian somatic follicle cells.
The class C synMuv genes encode components of a putative histone acetyltransferase complex similar to the human Tip60 and yeast NuA4 histone acetyltransferase complexes (Ceol and Horvitz 2004). The molecular identities of class B and class C synMuv genes suggest that chromatin remodeling and modification are important in specifying P(3–8).p fates. The class A synMuv genes lin-15A and lin-8 encode novel proteins (Clark et al. 1994; Huang et al. 1994; Davison et al. 2005). Little is known about the mechanism of action of the class A synMuv genes.
Previous synMuv genetic screens required that mutant isolates be fertile for the recovery of synMuv mutations. We used a screening approach that allowed the recovery of synMuv mutations that cause recessive sterility. We describe the characterization of new synMuv mutations and criteria used to distinguish new and previously described classes of synMuv genes.
MATERIALS AND METHODS
Strains and general techniques:
Strains were cultured as described by Brenner (1974) and grown at 20° unless otherwise indicated. The wild-type parent of all C. elegans strains described in this study was the Bristol strain N2, except that some multifactor mapping experiments used the polymorphic wild-type strains RW7000 (Williams et al. 1992) and CB4856 (Wicks et al. 2001). We also used strains containing the following mutations:
LGI: bli-3(e767), lin-17(n677), unc-11(e47), unc-73(e936), lin-44(n1792), unc-38(x20), dpy-5(e61), lin-35(n745), unc-13(e1091), lin-53(n833) (Ferguson and Horvitz 1989), and unc-54(e1092) (Dibb et al. 1985).
LGII: lin-31(n301), dpy-10(e128), tra-2(q276), rol-6(e187), dpl-1(n2994) (Ceol and Horvitz 2001; Thomas et al. 2003), let-23(sy10, sy97), unc-4(e120), unc-53(n569), mex-1(it9), rol-1(e91), and lin-38(n751).
LGIII: dpy-17(e164), lon-1(e185), lin-13(n770) (Ferguson and Horvitz 1989), lin-37(n758), lin-36(n766), unc-36(e251), lin-9(n112), unc-32(e189), lin-52(n771) (Ferguson and Horvitz 1989), and dpy-18(e364).
LGIV: lin-1(e1275), unc-5(e53), unc-24(e138), mec-3(e1338), lin-3(n378), sem-3(n1900) (M. J. Stern and H. R. Horvitz, unpublished results), dpy-20(e1282), unc-22(e66), dpy-26(n198), ark-1(sy247) (Hopper et al. 2000), unc-31(e169), unc-30(e191), lin-54(n2231) (Thomas et al. 2003), and dpy-4(e1166).
LGV: tam-1(cc567) (Hsieh et al. 1999), unc-46(e177), let-418(s1617), dpy-11(e224), rol-4(sc8), unc-76(e911), efl-1(n3318) (Ceol and Horvitz 2001), and dpy-21(e428).
LGX: egl-17(e1313), sli-1(sy143), aex-3(ad418), unc-1(e1598 n1201) (E. C. Park and H. R. Horvitz, unpublished results), dpy-3(e27), gap-1(ga133) (Hajnal et al. 1997), unc-2(e55), lon-2(e678), unc-10(e102), dpy-6(e14), unc-9(e101), unc-3(e151), lin-15B(n744), lin-15A(n767), and lin-15AB(n765).
Unless otherwise noted, the mutations used are described by Hodgkin (1997). In addition, we used strains containing the following chromosomal aberrations: mnDf57 II (Sigurdson et al. 1984), mnDf90 II (Sigurdson et al. 1984), mnDf29 II (Sigurdson et al. 1984), mnDf87 II (Sigurdson et al. 1984), mIn1[dpy-10(e128) mIs14] II (Edgley and Riddle 2001), mnC1[dpy-10(e128) unc-52(e444)] II (Herman 1978), nDf40 III (Hengartner et al. 1992), qC1[dpy-19(e1259) glp-1(q339)] III (Austin and Kimble, 1989), sDf63 IV (Clark and Baillie 1992), sDf62 IV (Clark and Baillie 1992), sDf10 IV (Rogalski et al. 1982), hT2[qIs48] (I;III) (L. Mathies and J. Kimble, personal communication), eT1(III;V) (Rosenbluth and Baillie 1981), nT1(IV;V) (Ferguson and Horvitz 1985), nT1(n754) (IV;V), and nT1[qIs51] (IV;V) (L. Mathies and J. Kimble, personal communication). n754 causes a dominant Unc phenotype, allowing nT1(n754)-containing larvae and adults to be scored (E. L. Ferguson and H. R. Horvitz, unpublished results). mIs14, an integrated transgene linked to the chromosomal inversion mIn1 (Edgley and Riddle 2001), and qIs48 and qIs51, integrated transgenes linked to the reciprocal translocations hT2(I;III) and nT1(IV;V), respectively (L. Mathies and J. Kimble, personal communication), consist of GFP-expressing transgenes that allow mIs14, qIs48, or qIs51-containing animals to be scored beginning at the four-cell stage of embryogenesis.
Isolation of new alleles:
We mutagenized lin-15A(n767) hermaphrodites with ethyl methanesulfonate (EMS) as described by Brenner (1974). We allowed these animals to recover on food for between 15 min and 1 hr and then transferred individual P0 larvae in L4 lethargus to 50-mm petri plates. After 3–5 days, 20 F1 L4 larvae per P0 were individually transferred to 50-mm plates, and F2 animals on these plates were subsequently screened for a Muv phenotype. We screened the progeny of 3380 F1 animals using this procedure.
Linkage group assignment:
We mapped newly isolated synMuv mutations to linkage groups using standard methods (Brenner 1974), except for some mutations that we mapped using the polymorphisms present in the wild-type strain RW7000 (Williams et al. 1992).
Complementation tests:
We performed complementation tests as described by Ferguson and Horvitz (1989). Hemizygous lin-15B(n3711) lin-15A(n767) males could not mate. To perform complementation tests with this mutation, we mated tra-2(q276); lin-15B(n3711) lin-15A(n767)/++ XX males with marked lin-15AB hermaphrodites and scored cross-progeny.
Construction of deficiency heterozygotes:
To construct trr-1(n3712) heterozygotes with the mnDf57, mnDf90, and mnDf29 deletions, Df/mIn1; lin-15A(n767) males were generated. These males were mated with rol-6 trr-1(n3712)/mIn1; lin-15A(n767) hermaphrodites, and non-Rol, non-Gfp cross-progeny were scored. mnDf87 heterozygous males do not mate, so in this case we generated trr-1(n3712)/mnDf87; lin-15A(n767) animals by mating trr-1(n3712)/mIn1; lin-15A(n767) males with unc-4 mnDf87/mIn1; lin-15A(n767) hermaphrodites. mep-1/Df animals were constructed by mating Df/nT1; +/nT1 males with dpy-20 mep-1; lin-15A(n767) hermaphrodites and scoring non-Dpy cross-progeny.
Construction of single-mutant and unlinked double-mutant strains:
The synMuv mutations listed below were balanced in trans by the specified double-mutant combinations or chromosomal aberrations in constructing strains with a single synMuv mutation or strains carrying two unlinked synMuv mutations:
lin-65(n3441): bli-3(e767) lin-17(n677), hT2[qIs48] (I;III).
lin(n3628): unc-11(e47) dpy-5(e61), hT2[qIs48] (I;III).
lin-35(n745): dpy-5(e61) unc-13(e1091), hT2[qIs48] (I;III).
trr-1(n3712): mIn1[dpy-10(e128) mIs14].
lin-38(n751): mnC1[dpy-10(e128) unc-52(e444)].
mep-1(n3703): dpy-20(e1282) unc-30(e191), nT1 n754 (IV;V), nT1[qIs51] (IV;V).
ark-1(n3701): dpy-20(e1282) unc-30(e191), nT1 n754 (IV;V), nT1[qIs51] (IV;V).
mys-1(n3681): unc-46(e177) dpy-11(e224), nT1 n754 (IV;V), nT1[qIs51] (IV;V).
let-23(sy97) was balanced with mIn1[dpy-10(e128) mIs14].
A sli-1 single mutant was constucted by generating + sli-1 + lin-15A/egl-17 + unc-1 + hermaphrodites and identifying nonmutant progeny that segregated only Egl Unc non-Muv and non-Egl non-Unc non-Muv animals. Non-Egl non-Unc non-Muv animals were isolated, and sli-1 homozygotes were identified as those that did not segregate Egl Unc non-Muv progeny. Double-mutant strains containing an X-linked mutation in sli-1, gap-1, lin-15A, or lin-15B and an autosomal mutation were constructed essentially as described by Ferguson and Horvitz (1989).
To ensure that mutations were not lost by recombination, several independent lines were isolated for each strain. Some double-mutant strains that exhibited a strong synMuv phenotype were constructed on the basis of their Muv phenotype without the use of balancers.
Construction of linked double-mutant strains:
To construct an n3628 lin-35 double mutant, hermaphrodites of genotype n3628 dpy-5 + +/+ + lin-35 unc-13; lin-15A were generated. Muv non-Dpy non-Unc progeny that segregated only Muv non-Dpy non-Unc, Muv Dpy non-Unc, and Muv Unc non-Dpy animals were selected. Muv Unc non-Dpy animals of the genotype n3628 lin-35 unc-13; lin-15A were isolated, and the lin-15A mutation was crossed out using unc-11 dpy-5 as a balancer.
To construct a sli-1 lin-15B double mutant, + sli-1 + lin-15A/egl-17 + unc-1 lin-15AB hermaphrodites were generated. Muv non-Egl non-Unc progeny that segregated only Muv non-Egl non-Unc and Muv Egl Unc animals were selected. Muv non-Egl non-Unc animals of the genotype sli-1 lin-15AB were isolated. From these animals, + sli-1 + lin-15AB/egl-17 + unc-1 lin-15B animals were generated. non-Muv non-Egl non-Unc progeny that segregated only non-Muv non-Egl non-Unc and Egl Unc non-Muv animals were identified, and non-Muv non-Egl non-Unc animals of the genotype sli-1 lin-15B were isolated. A gap-1 lin-15B double mutant was similarly constructed using dpy-3 unc-2 as a balancer.
A sli-1 gap-1 double mutant was constructed by generating sli-1 + dpy-3 +/+ unc-1 + gap-1 hermaphrodites and individually isolating non-Dpy non-Unc progeny. Progeny that segregated only non-Dpy non-Unc and Dpy non-Unc animals were identified, and non-Dpy non-Unc animals of the genotype sli-1 gap-1 were subsequently isolated.
Because trr-1(n3712) and let-23(sy97) cause recessive sterility and highly penetrant larval lethality, respectively, we could not isolate trr-1 or let-23 homozygotes in our construction of a trr-1 let-23 double mutant. For this reason, we built this double mutant by first generating + rol-6 + trr-1/let-23 + unc-4 +; lin-15A males and mating them with mIn1[dpy-10(e128) mIs14]; lin-15A hermaphrodites. Non-Dpy cross-progeny were individually isolated. Non-Dpy progeny with broods consisting of dead larvae and Vul Unc Gro non-Muv non-Rol non-Gfp and Gfp non-Vul non-Unc non-Gro non-Rol animals were identified. The presence of trr-1 in these broods, as judged by the trr-1-associated growth-rate abnormality (Gro), was later confirmed by complementation testing. lin-15A was crossed out to generate a let-23 unc-4 trr-1/mIn1[dpy-10(e128) mIs14] strain.
Assay for P(3–8).p vulval cell fates:
Cell fates were scored in L4 hermaphrodites using Nomarski microscopy by counting the number of descendants that had been produced by individual P(3–8).p cells. Scores of 1, 0.5, and 0 were assigned to cells that did fully, partially, or not adopt vulval cell fates, respectively. P(3–8).p cells that partially adopt a vulval cell fate have one daughter that divides to produce two to four descendants and another daughter that remains undivided (Aroian and Sternberg 1991).
RNA-mediated interference:
Templates for in vitro transcription reactions were made by PCR amplification of cDNAs and their flanking T3 and T7 promoter sequences. In vitro transcribed RNA was denatured for 10 min and subsequently annealed prior to injection.
lin-65 rescue:
Using Gateway in vitro recombination technology (Invitrogen, San Diego), we cloned the open reading frame encoding the 728-amino-acid LIN-65 protein from a pENTR201lin-65 entry clone into the pMB1 and pMB7 destination vectors. pMB1 and pMB7 (kindly provided by M. Boxem and S. van den Heuvel) are designed to express inserted sequences under the control of the C. elegans heat-shock protein promoters Phsp16-2 and Phsp16-41, respectively. We performed transformation rescue (Mello et al. 1991) using the green fluorescent protein-expressing plasmid pTG96 (kindly provided by Min Han) as a coinjection marker. Transgenic animals were heat-shocked as L1 and L2 larvae for 1 hr at 33° and scored as adults. Control transgenic animals were not heat-shocked.
Allele sequence:
We used PCR-amplified regions of genomic DNA as templates in determining gene sequences. For each gene investigated, we determined the sequences of all exons and splice junctions. Whenever observed, the sequence of a mutation was confirmed using an independently derived PCR product. All sequences were determined using an automated ABI 373 DNA sequencer (Applied Biosystems, Foster City, CA).
RESULTS
Isolation of new synMuv mutants:
A severe reduction of class B synMuv gene function is often associated with sterility: (1) In a genetic screen for alleles that did not complement the synMuv phenotype of lin-9(n112), Ferguson and Horvitz (1989) recovered two lin-9 alleles, n942 and n943, that caused recessive sterility; (2) gene-dosage studies indicate that, in comparison to the wild type, lin-52(n771)/Df and dpl-1(n2994)/Df heterozygotes have markedly reduced brood sizes (Ceol and Horvitz 2001; Thomas et al. 2003); and (3) deletion mutations of some synMuv genes recovered using a PCR-based screening approach show recessive sterility, e.g., mutations of lin-53 (Lu 1999), efl-1, and dpl-1 (Ceol and Horvitz 2001).
Previous genetic screens for synMuv mutants (Ferguson and Horvitz 1989; Thomas et al. 2003) were performed before a connection between loss of synMuv gene function and sterility was well established. These screens required that isolates be fertile and viable for the recovery of mutant alleles and failed to recover mutations of the class B synMuv genes efl-1 and let-418, both of which can mutate to cause a sterile phenotype (von Zelewsky et al. 2000; Ceol and Horvitz 2001). These results suggested that additional synMuv genes might be identified in a screen that allowed the recovery of homozygous sterile mutations.
To screen for new synMuv mutants, we examined the progeny of individual F1 animals after EMS mutagenesis of their lin-15A(n767) parents. We screened the progeny of 3380 F1 animals (6760 haploid genomes) for mutations that either alone or in combination with lin-15A(n767) caused a recessive Muv phenotype. Mutations that caused recessive sterility in addition to a Muv phenotype were recovered from their heterozygous wild-type siblings present on the same petri plate. Using this strategy we identified 95 Muv mutations, 24 of which we maintained as heterozygotes because of a recessive sterility that cosegregated with the Muv phenotype. Three mutations caused a Muv phenotype in the absence of lin-15A(n767) and were found to affect the previously studied genes lin-1 and lin-31, both of which function downstream of let-60 Ras in vulval induction (Ferguson et al. 1987). These mutations, lin-1(n3443), lin-1(n3522), and lin-31(n3440), were not characterized further. Thirty mutations when in combination with lin-15A(n767) caused a weakly penetrant (<30%) Muv phenotype. We were unable to convincingly map these mutations to linkage groups. The remaining 62 mutations were assigned to 20 complementation groups (see below). Five of these mutations affect the synMuv gene lin-61 and will be described elsewhere (M. M. Harrison, X. Lu and H. R. Horvitz, unpublished results).
Phenotypes of new mutants:
We characterized the penetrance of the Muv phenotype of each strain at 15° and 20° (Table 1
Phenotypes of synMuv mutant strains
. | % Muv (n) . | . | |
|---|---|---|---|
| Genotype . | 15° . | 20° . | Additional abnormalities . |
| ark-1(n3524); lin-15A(n767) | 0 (251) | 80 (171) | |
| ark-1(n3701); lin-15A(n767) | 12 (190) | 95 (160) | |
| dpl-1(n3643); lin-15A(n767)a | 99 (154) | 100 (252) | |
| efl-1(n3639); lin-15A(n767)a | 93 (74) | 100 (78) | Ste |
| gap-1(n3535) lin-15A(n767) | 1 (143) | 50 (236) | |
| let-418(n3536); lin-15A(n767) | 0 (201) | 55 (183) | hs Ste |
| let-418(n3626); lin-15A(n767) | 2 (62) | 97 (76) | Ste |
| let-418(n3629); lin-15A(n767) | 0 (52) | 86 (58) | Ste |
| let-418(n3634); lin-15A(n767) | 0 (87) | 92 (48) | Ste |
| let-418(n3635); lin-15A(n767) | 0 (76) | 71 (70) | Ste |
| let-418(n3636); lin-15A(n767) | 0 (77) | 92 (78) | Ste |
| let-418(n3719); lin-15A(n767) | 0 (101) | 100 (60) | Ste |
| lin-9(n3631); lin-15A(n767) | 100 (42) | 100 (72) | Ste |
| lin-9(n3675); lin-15A(n767) | 43 (166) | 100 (105) | |
| lin-9(n3767); lin-15A(n767) | 100 (67) | 100 (56) | Ste |
| lin-13(n3642); lin-15A(n767) | 3 (60) | 100 (63) | Ste |
| lin-13(n3673); lin-15A(n767) | 61 (145) | 97 (129) | |
| lin-13(n3674); lin-15A(n767) | 78 (131) | 100 (191) | hs Ste |
| lin-13(n3726); lin-15A(n767) | 31 (225) | 99 (149) | hs Ste |
| lin-15B(n3436) lin-15A(n767) | 100 (193) | 100 (212) | |
| lin-15B(n3676) lin-15A(n767) | 18 (167) | 72 (130) | |
| lin-15B(n3677) lin-15A(n767) | 99 (111) | 100 (122) | |
| lin-15B(n3711) lin-15A(n767) | 100 (186) | 100 (156) | |
| lin-15B(n3760) lin-15A(n767) | 32 (171) | 100 (150) | |
| lin-15B(n3762) lin-15A(n767) | 63 (113) | 97 (116) | |
| lin-15B(n3764) lin-15A(n767) | 96 (232) | 100 (199) | |
| lin-15B(n3766) lin-15A(n767) | 55 (132) | 100 (173) | |
| lin-15B(n3768) lin-15A(n767) | 80 (159) | 100 (302) | |
| lin-15B(n3772) lin-15A(n767) | 100 (220) | 100 (191) | |
| lin-35(n3438); lin-15A(n767) | 100 (153) | 100 (126) | Partial Ste at 20°, Rup |
| lin-35(n3763); lin-15A(n767) | 100 (108) | 100 (160) | Partial Ste at 20°, Rup |
| lin-36(n3671); lin-15A(n767) | 65 (191) | 100 (151) | |
| lin-36(n3672); lin-15A(n767) | 98 (198) | 100 (178) | |
| lin-36(n3765); lin-15A(n767) | 0 (184) | 37 (202) | |
| lin-52(n3718); lin-15A(n767)b | 100 (41) | 100 (82) | Ste |
| lin-53(n3448); lin-15A(n767) | 67 (130) | 100 (211) | Partial Ste at 20° |
| lin-53(n3521); lin-15A(n767) | 100 (34) | 100 (125) | Partial Ste at 20° |
| lin-53(n3622); lin-15A(n767) | 85 (61) | 100 (66) | Ste |
| lin-53(n3623); lin-15A(n767) | 24 (55) | 100 (51) | Ste |
| lin-65(n3441); lin-15A(n767) | 80 (165) | 99 (195) | |
| lin-65(n3541); lin-15A(n767) | 79 (242) | 98 (137) | |
| lin-65(n3543); lin-15A(n767) | 85 (177) | 100 (121) | |
| lin(n3628); lin-15A(n767) | 3 (103) | 84 (188) | |
| lin(n3542) lin-15A(n767) | 0 (127) | 35 (218) | |
| mep-1(n3680); lin-15A(n767) | 5 (122) | 97 (105) | hs Ste |
| mep-1(n3702); lin-15A(n767) | 30 (61) | 100 (141) | Ste |
| mep-1(n3703); lin-15A(n767) | 25 (72) | 100 (107) | Ste |
| mys-1(n3681); lin-15A(n767)c | 0 (214) | 72 (192) | |
| sli-1(n3538) lin-15A(n767) | 4 (138) | 90 (173) | |
| sli-1(n3544) lin-15A(n767) | 5 (153) | 80 (265) | cs embryonic lethality |
| sli-1(n3683) lin-15A(n767) | 5 (80) | 88 (148) | cs embryonic lethality |
| trr-1(n3630); lin-15A(n767)c | 3 (131) | 85 (212) | Ste, Gro |
| trr-1(n3637); lin-15A(n767)c | 1 (92) | 80 (200) | Ste, Gro |
| trr-1(n3704); lin-15A(n767)c | 3 (96) | 79 (244) | Ste, Gro |
| trr-1(n3708); lin-15A(n767)c | 2 (151) | 84 (228) | Ste, Gro |
| trr-1(n3709); lin-15A(n767)c | 1 (97) | 77 (154) | Ste, Gro |
| trr-1(n3712); lin-15A(n767)c | 6 (121) | 77 (192) | Ste, Gro |
. | % Muv (n) . | . | |
|---|---|---|---|
| Genotype . | 15° . | 20° . | Additional abnormalities . |
| ark-1(n3524); lin-15A(n767) | 0 (251) | 80 (171) | |
| ark-1(n3701); lin-15A(n767) | 12 (190) | 95 (160) | |
| dpl-1(n3643); lin-15A(n767)a | 99 (154) | 100 (252) | |
| efl-1(n3639); lin-15A(n767)a | 93 (74) | 100 (78) | Ste |
| gap-1(n3535) lin-15A(n767) | 1 (143) | 50 (236) | |
| let-418(n3536); lin-15A(n767) | 0 (201) | 55 (183) | hs Ste |
| let-418(n3626); lin-15A(n767) | 2 (62) | 97 (76) | Ste |
| let-418(n3629); lin-15A(n767) | 0 (52) | 86 (58) | Ste |
| let-418(n3634); lin-15A(n767) | 0 (87) | 92 (48) | Ste |
| let-418(n3635); lin-15A(n767) | 0 (76) | 71 (70) | Ste |
| let-418(n3636); lin-15A(n767) | 0 (77) | 92 (78) | Ste |
| let-418(n3719); lin-15A(n767) | 0 (101) | 100 (60) | Ste |
| lin-9(n3631); lin-15A(n767) | 100 (42) | 100 (72) | Ste |
| lin-9(n3675); lin-15A(n767) | 43 (166) | 100 (105) | |
| lin-9(n3767); lin-15A(n767) | 100 (67) | 100 (56) | Ste |
| lin-13(n3642); lin-15A(n767) | 3 (60) | 100 (63) | Ste |
| lin-13(n3673); lin-15A(n767) | 61 (145) | 97 (129) | |
| lin-13(n3674); lin-15A(n767) | 78 (131) | 100 (191) | hs Ste |
| lin-13(n3726); lin-15A(n767) | 31 (225) | 99 (149) | hs Ste |
| lin-15B(n3436) lin-15A(n767) | 100 (193) | 100 (212) | |
| lin-15B(n3676) lin-15A(n767) | 18 (167) | 72 (130) | |
| lin-15B(n3677) lin-15A(n767) | 99 (111) | 100 (122) | |
| lin-15B(n3711) lin-15A(n767) | 100 (186) | 100 (156) | |
| lin-15B(n3760) lin-15A(n767) | 32 (171) | 100 (150) | |
| lin-15B(n3762) lin-15A(n767) | 63 (113) | 97 (116) | |
| lin-15B(n3764) lin-15A(n767) | 96 (232) | 100 (199) | |
| lin-15B(n3766) lin-15A(n767) | 55 (132) | 100 (173) | |
| lin-15B(n3768) lin-15A(n767) | 80 (159) | 100 (302) | |
| lin-15B(n3772) lin-15A(n767) | 100 (220) | 100 (191) | |
| lin-35(n3438); lin-15A(n767) | 100 (153) | 100 (126) | Partial Ste at 20°, Rup |
| lin-35(n3763); lin-15A(n767) | 100 (108) | 100 (160) | Partial Ste at 20°, Rup |
| lin-36(n3671); lin-15A(n767) | 65 (191) | 100 (151) | |
| lin-36(n3672); lin-15A(n767) | 98 (198) | 100 (178) | |
| lin-36(n3765); lin-15A(n767) | 0 (184) | 37 (202) | |
| lin-52(n3718); lin-15A(n767)b | 100 (41) | 100 (82) | Ste |
| lin-53(n3448); lin-15A(n767) | 67 (130) | 100 (211) | Partial Ste at 20° |
| lin-53(n3521); lin-15A(n767) | 100 (34) | 100 (125) | Partial Ste at 20° |
| lin-53(n3622); lin-15A(n767) | 85 (61) | 100 (66) | Ste |
| lin-53(n3623); lin-15A(n767) | 24 (55) | 100 (51) | Ste |
| lin-65(n3441); lin-15A(n767) | 80 (165) | 99 (195) | |
| lin-65(n3541); lin-15A(n767) | 79 (242) | 98 (137) | |
| lin-65(n3543); lin-15A(n767) | 85 (177) | 100 (121) | |
| lin(n3628); lin-15A(n767) | 3 (103) | 84 (188) | |
| lin(n3542) lin-15A(n767) | 0 (127) | 35 (218) | |
| mep-1(n3680); lin-15A(n767) | 5 (122) | 97 (105) | hs Ste |
| mep-1(n3702); lin-15A(n767) | 30 (61) | 100 (141) | Ste |
| mep-1(n3703); lin-15A(n767) | 25 (72) | 100 (107) | Ste |
| mys-1(n3681); lin-15A(n767)c | 0 (214) | 72 (192) | |
| sli-1(n3538) lin-15A(n767) | 4 (138) | 90 (173) | |
| sli-1(n3544) lin-15A(n767) | 5 (153) | 80 (265) | cs embryonic lethality |
| sli-1(n3683) lin-15A(n767) | 5 (80) | 88 (148) | cs embryonic lethality |
| trr-1(n3630); lin-15A(n767)c | 3 (131) | 85 (212) | Ste, Gro |
| trr-1(n3637); lin-15A(n767)c | 1 (92) | 80 (200) | Ste, Gro |
| trr-1(n3704); lin-15A(n767)c | 3 (96) | 79 (244) | Ste, Gro |
| trr-1(n3708); lin-15A(n767)c | 2 (151) | 84 (228) | Ste, Gro |
| trr-1(n3709); lin-15A(n767)c | 1 (97) | 77 (154) | Ste, Gro |
| trr-1(n3712); lin-15A(n767)c | 6 (121) | 77 (192) | Ste, Gro |
The penetrance of the Muv phenotype was determined after synMuv mutant strains grew at the indicated temperature for two or more generations. For most strains for which a fully penetrant sterile phenotype was associated with the Muv phenotype, we scored the penetrance of the Muv phenotype by examining sterile progeny of heterozygous mutant parents. For trr-1 mutant strains, we scored the penetrance of the Muv phenotype by examining non-Gfp progeny of trr-1/mIn1[dpy-10(e128)mIs14]; lin-15A(n767) heterozygous parents. All strains were backcrossed to lin-15A(n767) twice prior to phenotypic characterization. In addition to the phenotypes described above, many of the strains exhibited heat-sensitive inviability as a consequence of rupture and/or general sickness. Ste, sterile; Gro, growth rate abnormal; Rup, rupture at the vulva; cs, cold sensitive; hs, heat sensitive. The characterization of some of these strains was previously described by:
Phenotypes of synMuv mutant strains
. | % Muv (n) . | . | |
|---|---|---|---|
| Genotype . | 15° . | 20° . | Additional abnormalities . |
| ark-1(n3524); lin-15A(n767) | 0 (251) | 80 (171) | |
| ark-1(n3701); lin-15A(n767) | 12 (190) | 95 (160) | |
| dpl-1(n3643); lin-15A(n767)a | 99 (154) | 100 (252) | |
| efl-1(n3639); lin-15A(n767)a | 93 (74) | 100 (78) | Ste |
| gap-1(n3535) lin-15A(n767) | 1 (143) | 50 (236) | |
| let-418(n3536); lin-15A(n767) | 0 (201) | 55 (183) | hs Ste |
| let-418(n3626); lin-15A(n767) | 2 (62) | 97 (76) | Ste |
| let-418(n3629); lin-15A(n767) | 0 (52) | 86 (58) | Ste |
| let-418(n3634); lin-15A(n767) | 0 (87) | 92 (48) | Ste |
| let-418(n3635); lin-15A(n767) | 0 (76) | 71 (70) | Ste |
| let-418(n3636); lin-15A(n767) | 0 (77) | 92 (78) | Ste |
| let-418(n3719); lin-15A(n767) | 0 (101) | 100 (60) | Ste |
| lin-9(n3631); lin-15A(n767) | 100 (42) | 100 (72) | Ste |
| lin-9(n3675); lin-15A(n767) | 43 (166) | 100 (105) | |
| lin-9(n3767); lin-15A(n767) | 100 (67) | 100 (56) | Ste |
| lin-13(n3642); lin-15A(n767) | 3 (60) | 100 (63) | Ste |
| lin-13(n3673); lin-15A(n767) | 61 (145) | 97 (129) | |
| lin-13(n3674); lin-15A(n767) | 78 (131) | 100 (191) | hs Ste |
| lin-13(n3726); lin-15A(n767) | 31 (225) | 99 (149) | hs Ste |
| lin-15B(n3436) lin-15A(n767) | 100 (193) | 100 (212) | |
| lin-15B(n3676) lin-15A(n767) | 18 (167) | 72 (130) | |
| lin-15B(n3677) lin-15A(n767) | 99 (111) | 100 (122) | |
| lin-15B(n3711) lin-15A(n767) | 100 (186) | 100 (156) | |
| lin-15B(n3760) lin-15A(n767) | 32 (171) | 100 (150) | |
| lin-15B(n3762) lin-15A(n767) | 63 (113) | 97 (116) | |
| lin-15B(n3764) lin-15A(n767) | 96 (232) | 100 (199) | |
| lin-15B(n3766) lin-15A(n767) | 55 (132) | 100 (173) | |
| lin-15B(n3768) lin-15A(n767) | 80 (159) | 100 (302) | |
| lin-15B(n3772) lin-15A(n767) | 100 (220) | 100 (191) | |
| lin-35(n3438); lin-15A(n767) | 100 (153) | 100 (126) | Partial Ste at 20°, Rup |
| lin-35(n3763); lin-15A(n767) | 100 (108) | 100 (160) | Partial Ste at 20°, Rup |
| lin-36(n3671); lin-15A(n767) | 65 (191) | 100 (151) | |
| lin-36(n3672); lin-15A(n767) | 98 (198) | 100 (178) | |
| lin-36(n3765); lin-15A(n767) | 0 (184) | 37 (202) | |
| lin-52(n3718); lin-15A(n767)b | 100 (41) | 100 (82) | Ste |
| lin-53(n3448); lin-15A(n767) | 67 (130) | 100 (211) | Partial Ste at 20° |
| lin-53(n3521); lin-15A(n767) | 100 (34) | 100 (125) | Partial Ste at 20° |
| lin-53(n3622); lin-15A(n767) | 85 (61) | 100 (66) | Ste |
| lin-53(n3623); lin-15A(n767) | 24 (55) | 100 (51) | Ste |
| lin-65(n3441); lin-15A(n767) | 80 (165) | 99 (195) | |
| lin-65(n3541); lin-15A(n767) | 79 (242) | 98 (137) | |
| lin-65(n3543); lin-15A(n767) | 85 (177) | 100 (121) | |
| lin(n3628); lin-15A(n767) | 3 (103) | 84 (188) | |
| lin(n3542) lin-15A(n767) | 0 (127) | 35 (218) | |
| mep-1(n3680); lin-15A(n767) | 5 (122) | 97 (105) | hs Ste |
| mep-1(n3702); lin-15A(n767) | 30 (61) | 100 (141) | Ste |
| mep-1(n3703); lin-15A(n767) | 25 (72) | 100 (107) | Ste |
| mys-1(n3681); lin-15A(n767)c | 0 (214) | 72 (192) | |
| sli-1(n3538) lin-15A(n767) | 4 (138) | 90 (173) | |
| sli-1(n3544) lin-15A(n767) | 5 (153) | 80 (265) | cs embryonic lethality |
| sli-1(n3683) lin-15A(n767) | 5 (80) | 88 (148) | cs embryonic lethality |
| trr-1(n3630); lin-15A(n767)c | 3 (131) | 85 (212) | Ste, Gro |
| trr-1(n3637); lin-15A(n767)c | 1 (92) | 80 (200) | Ste, Gro |
| trr-1(n3704); lin-15A(n767)c | 3 (96) | 79 (244) | Ste, Gro |
| trr-1(n3708); lin-15A(n767)c | 2 (151) | 84 (228) | Ste, Gro |
| trr-1(n3709); lin-15A(n767)c | 1 (97) | 77 (154) | Ste, Gro |
| trr-1(n3712); lin-15A(n767)c | 6 (121) | 77 (192) | Ste, Gro |
. | % Muv (n) . | . | |
|---|---|---|---|
| Genotype . | 15° . | 20° . | Additional abnormalities . |
| ark-1(n3524); lin-15A(n767) | 0 (251) | 80 (171) | |
| ark-1(n3701); lin-15A(n767) | 12 (190) | 95 (160) | |
| dpl-1(n3643); lin-15A(n767)a | 99 (154) | 100 (252) | |
| efl-1(n3639); lin-15A(n767)a | 93 (74) | 100 (78) | Ste |
| gap-1(n3535) lin-15A(n767) | 1 (143) | 50 (236) | |
| let-418(n3536); lin-15A(n767) | 0 (201) | 55 (183) | hs Ste |
| let-418(n3626); lin-15A(n767) | 2 (62) | 97 (76) | Ste |
| let-418(n3629); lin-15A(n767) | 0 (52) | 86 (58) | Ste |
| let-418(n3634); lin-15A(n767) | 0 (87) | 92 (48) | Ste |
| let-418(n3635); lin-15A(n767) | 0 (76) | 71 (70) | Ste |
| let-418(n3636); lin-15A(n767) | 0 (77) | 92 (78) | Ste |
| let-418(n3719); lin-15A(n767) | 0 (101) | 100 (60) | Ste |
| lin-9(n3631); lin-15A(n767) | 100 (42) | 100 (72) | Ste |
| lin-9(n3675); lin-15A(n767) | 43 (166) | 100 (105) | |
| lin-9(n3767); lin-15A(n767) | 100 (67) | 100 (56) | Ste |
| lin-13(n3642); lin-15A(n767) | 3 (60) | 100 (63) | Ste |
| lin-13(n3673); lin-15A(n767) | 61 (145) | 97 (129) | |
| lin-13(n3674); lin-15A(n767) | 78 (131) | 100 (191) | hs Ste |
| lin-13(n3726); lin-15A(n767) | 31 (225) | 99 (149) | hs Ste |
| lin-15B(n3436) lin-15A(n767) | 100 (193) | 100 (212) | |
| lin-15B(n3676) lin-15A(n767) | 18 (167) | 72 (130) | |
| lin-15B(n3677) lin-15A(n767) | 99 (111) | 100 (122) | |
| lin-15B(n3711) lin-15A(n767) | 100 (186) | 100 (156) | |
| lin-15B(n3760) lin-15A(n767) | 32 (171) | 100 (150) | |
| lin-15B(n3762) lin-15A(n767) | 63 (113) | 97 (116) | |
| lin-15B(n3764) lin-15A(n767) | 96 (232) | 100 (199) | |
| lin-15B(n3766) lin-15A(n767) | 55 (132) | 100 (173) | |
| lin-15B(n3768) lin-15A(n767) | 80 (159) | 100 (302) | |
| lin-15B(n3772) lin-15A(n767) | 100 (220) | 100 (191) | |
| lin-35(n3438); lin-15A(n767) | 100 (153) | 100 (126) | Partial Ste at 20°, Rup |
| lin-35(n3763); lin-15A(n767) | 100 (108) | 100 (160) | Partial Ste at 20°, Rup |
| lin-36(n3671); lin-15A(n767) | 65 (191) | 100 (151) | |
| lin-36(n3672); lin-15A(n767) | 98 (198) | 100 (178) | |
| lin-36(n3765); lin-15A(n767) | 0 (184) | 37 (202) | |
| lin-52(n3718); lin-15A(n767)b | 100 (41) | 100 (82) | Ste |
| lin-53(n3448); lin-15A(n767) | 67 (130) | 100 (211) | Partial Ste at 20° |
| lin-53(n3521); lin-15A(n767) | 100 (34) | 100 (125) | Partial Ste at 20° |
| lin-53(n3622); lin-15A(n767) | 85 (61) | 100 (66) | Ste |
| lin-53(n3623); lin-15A(n767) | 24 (55) | 100 (51) | Ste |
| lin-65(n3441); lin-15A(n767) | 80 (165) | 99 (195) | |
| lin-65(n3541); lin-15A(n767) | 79 (242) | 98 (137) | |
| lin-65(n3543); lin-15A(n767) | 85 (177) | 100 (121) | |
| lin(n3628); lin-15A(n767) | 3 (103) | 84 (188) | |
| lin(n3542) lin-15A(n767) | 0 (127) | 35 (218) | |
| mep-1(n3680); lin-15A(n767) | 5 (122) | 97 (105) | hs Ste |
| mep-1(n3702); lin-15A(n767) | 30 (61) | 100 (141) | Ste |
| mep-1(n3703); lin-15A(n767) | 25 (72) | 100 (107) | Ste |
| mys-1(n3681); lin-15A(n767)c | 0 (214) | 72 (192) | |
| sli-1(n3538) lin-15A(n767) | 4 (138) | 90 (173) | |
| sli-1(n3544) lin-15A(n767) | 5 (153) | 80 (265) | cs embryonic lethality |
| sli-1(n3683) lin-15A(n767) | 5 (80) | 88 (148) | cs embryonic lethality |
| trr-1(n3630); lin-15A(n767)c | 3 (131) | 85 (212) | Ste, Gro |
| trr-1(n3637); lin-15A(n767)c | 1 (92) | 80 (200) | Ste, Gro |
| trr-1(n3704); lin-15A(n767)c | 3 (96) | 79 (244) | Ste, Gro |
| trr-1(n3708); lin-15A(n767)c | 2 (151) | 84 (228) | Ste, Gro |
| trr-1(n3709); lin-15A(n767)c | 1 (97) | 77 (154) | Ste, Gro |
| trr-1(n3712); lin-15A(n767)c | 6 (121) | 77 (192) | Ste, Gro |
The penetrance of the Muv phenotype was determined after synMuv mutant strains grew at the indicated temperature for two or more generations. For most strains for which a fully penetrant sterile phenotype was associated with the Muv phenotype, we scored the penetrance of the Muv phenotype by examining sterile progeny of heterozygous mutant parents. For trr-1 mutant strains, we scored the penetrance of the Muv phenotype by examining non-Gfp progeny of trr-1/mIn1[dpy-10(e128)mIs14]; lin-15A(n767) heterozygous parents. All strains were backcrossed to lin-15A(n767) twice prior to phenotypic characterization. In addition to the phenotypes described above, many of the strains exhibited heat-sensitive inviability as a consequence of rupture and/or general sickness. Ste, sterile; Gro, growth rate abnormal; Rup, rupture at the vulva; cs, cold sensitive; hs, heat sensitive. The characterization of some of these strains was previously described by:
). At 25° the penetrance of each strain was between 98 and 100% (n ≥ 25), except for gap-1(n3535); lin-15A(n767) (91%, n = 111) and lin(n3542) lin-15A(n767) (90%, n = 42). Since a heat-sensitive Muv phenotype is characteristic of most synMuv strains, including those with null mutations in synMuv genes, it is likely that many individual synMuv mutations are not temperature sensitive but rather that the synMuv genes regulate a temperature-sensitive process (Ferguson and Horvitz 1989).
As described in Table 1, many of these synMuv strains also exhibited a sterile phenotype. In these strains, the sterile phenotype cosegregated with the Muv phenotype during backcrosses and two- and three-factor mapping experiments. For efl-1, let-418, and certain lin-9 and lin-53 mutations, we found that our new mutations did not complement the sterile phenotypes caused by previously isolated allelic synMuv mutations (data not shown). Mutations defining new synMuv loci likewise failed to complement each other for the sterile phenotype: mep-1(n3702) did not complement mep-1(n3703), and none of the other five trr-1 mutations complemented trr-1(n3712) for the sterile phenotype. These observations indicate that the sterile and Muv phenotypes of these strains were caused by the same mutation.
New synMuv genes:
Using two-factor crosses and X chromosome transmission tests (see materials and methods), we mapped the new mutations to linkage groups. We then determined if each mutation failed to complement mutations in known synMuv genes on the same linkage group. In these tests we identified 41 alleles of known synMuv genes: 1 dpl-1, 1 efl-1, 7 let-418, 3 lin-9, 4 lin-13, 10 lin-15B, 2 lin-35, 3 lin-36, 1 lin-52, 4 lin-53, and 5 lin-61 mutations. We isolated 1 mutation in gap-1 and 3 in sli-1, two genes that were originally identified in screens for mutations that suppress the Vul phenotype caused by a reduction in let-60 Ras pathway signaling (Jongeward et al. 1995; Hajnal et al. 1997). We also identified two mutations in ark-1, a gene first identified in a screen for mutations that cause ectopic vulval cell fates in a sli-1 mutant background (Hopper et al. 2000). gap-1, sli-1, and ark-1 single mutants were previously found to have no (sli-1, gap-1) or subtle (ark-1) defects in vulval development. Our results indicate that sli-1, gap-1, and ark-1 act redundantly with lin-15A to negatively regulate let-60 Ras signaling.
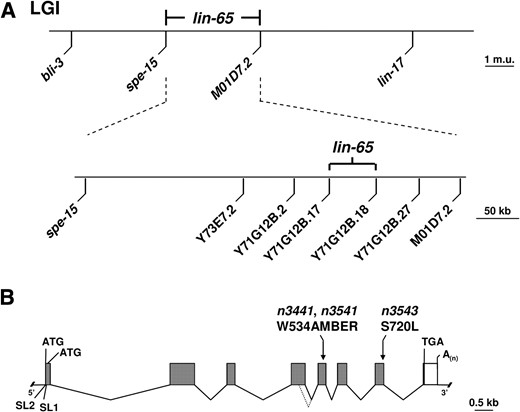
Mutations that were not assigned to known synMuv complementation groups were tested against unassigned mutations on the same linkage group for complementation. These tests defined five new synMuv loci: lin-65, lin(n3628), mep-1, mys-1, and trr-1. [lin(n3542) may define another new synMuv locus, but since we have not separated lin(n3542) from lin-15A(n767), we do not know whether lin(n3542) is a synMuv mutation or whether it causes a Muv phenotype on its own.] We used multifactor crosses (Table 2
Three- and four-factor crosses
Gene . | Genotype of heterozygote . | Phenotype of selected recombinants . | Genotype of selected recombinants (with respect to unselected markers) . |
|---|---|---|---|
| lin-65 | + lin-65 +/bli-3 + lin-17; lin-15A(n767) | Lin-17 | 9/19 lin-65/+ |
| bli-3 + lin-65/+ spe-15 +; lin-15A(n767) | Muv | 10/18 spe-15/+ | |
| + lin-65 lin-17/spe-15 + +; lin-15A(n767) | Lin-17 | 11/11 spe-15/+ | |
| bli-3 + + + lin-65 +/+ Y73E7.2 Y71G12B.2 Y71G12B.17 + Y71G12B.18; lin-15A(n767) | Muv | 4/30 Y73E7.2/+ | |
| Muv | 2/30 Y71G12B.2/+ | ||
| Muv | 1/30 Y71G12B.17/+ | ||
| Muv | 0/30 Y71G12B.18/+ | ||
| + lin-65 + + + lin-17/Y71G12B.17 + Y71G12B.18 Y71G12B.27 M01D7.2 +; lin-15A(n767) | Lin-17 | 17/23 M01D7.2/+ | |
| Lin-17 | 18/23 Y71G12B.27/+ | ||
| Lin-17 | 21/23 Y71G12B.18/+ | ||
| Lin-17 | 23/23 Y71G12B.17/+ | ||
| lin(n3542) | + + lin(n3542) lin-15A(n767)/unc-10 dpy-6 + lin-15A(n767) | Unc | 8/8 lin(n3542)/+ |
| + lin(n3542) + lin-15A(n767)/dpy-6 + unc-9 lin-15A(n767) | Unc | 4/40 lin(n3542)/+ | |
| lin(n3628) | lin(n3628) + +/+ dpy-5 unc-13; lin-15A(n767) | Dpy | 0/6 lin(n3628)/+ |
| Unc | 6/6 lin(n3628)/+ | ||
| + lin(n3628) +/unc-11 + dpy-5; lin-15A(n767) | Unc | 1/11 lin(n3628)/+ | |
| Dpy | 5/11 lin(n3628)/+ | ||
| unc-11 + + lin(n3628)/+ unc-73 lin-44 +; lin-15A(n767) | Muv | 3/9 unc-73 lin-44/++ | |
| + + lin(n3628) dpy-5/unc-73 lin-44 + +; lin-15A(n767) | Muv | 0/21 unc-73 lin-44/++ | |
| lin(n3628) + dpy-5/+ unc-38 +; lin-15A(n767) | Muv | 3/7 unc-38/+ | |
| unc-11 lin(n3628) +/+ + unc-38; lin-15A(n767) | Muv | 0/9 unc-38/+ | |
| mep-1 | + mep-1 +/unc-5 + dpy-20; lin-15A(n767) | Unc | 56/57 mep-1/+ |
| Dpy | 2/61 mep-1/+ | ||
| mep-1 + +/+ dpy-20 unc-30; lin-15A(n767) | Dpy | 0/51 mep-1/+ | |
| Unc | 58/58 mep-1/+ | ||
| + + mep-1 +/unc-24 mec-3 + dpy-20; lin-15A(n767) | Unc Mec | 10/12 mep-1/+ | |
| Unc | 17/17 mep-1/+ | ||
| Mec Dpy | 0/8 mep-1/+ | ||
| Dpy | 2/8 mep-1/+ | ||
| + mep-1 dpy-20 +/lin-3 + + unc-22; lin-15A(n767) | Dpy | 5/5 lin-3/+ | |
| Vul | 3/10 mep-1/+ | ||
| + + mep-1+/mec-3 sem-3 + dpy-20; lin-15A(n767) | Mec | 17/17 mep-1/+ | |
| Dpy | 6/13 mep-1/+ | ||
| mys-1 | + mys-1 +/unc-46 + dpy-11; lin-15A | Unc | 3/7 mys-1/+ |
| Dpy | 7/11 mys-1/+ | ||
| trr-1 | + rol-6 + trr-1/dpy-10 + unc-4 +; lin-15A(n767) | Rol | 3/14 unc-4/+ |
| Dpy | 3/3 trr-1/+ | ||
| Unc | 0/8 trr-1/+ | ||
| + trr-1 +/dpy-10 + rol-1; lin-15A(n767) | Rol | 9/20 trr-1/+ | |
| + + trr-1/dpy-10 unc-53 +; lin-15A(n767) | Unc | 0/17 trr-1/+ | |
| + trr-1 +/unc-53 + rol-1; lin-15A(n767) | Unc | 7/10 trr-1/+ | |
| Rol | 7/10 trr-1/+ | ||
| + trr-1 + rol-1/unc-4 + mex-1 +; lin-15A(n767) | Rol | 12/14 mex-1/+ |
Gene . | Genotype of heterozygote . | Phenotype of selected recombinants . | Genotype of selected recombinants (with respect to unselected markers) . |
|---|---|---|---|
| lin-65 | + lin-65 +/bli-3 + lin-17; lin-15A(n767) | Lin-17 | 9/19 lin-65/+ |
| bli-3 + lin-65/+ spe-15 +; lin-15A(n767) | Muv | 10/18 spe-15/+ | |
| + lin-65 lin-17/spe-15 + +; lin-15A(n767) | Lin-17 | 11/11 spe-15/+ | |
| bli-3 + + + lin-65 +/+ Y73E7.2 Y71G12B.2 Y71G12B.17 + Y71G12B.18; lin-15A(n767) | Muv | 4/30 Y73E7.2/+ | |
| Muv | 2/30 Y71G12B.2/+ | ||
| Muv | 1/30 Y71G12B.17/+ | ||
| Muv | 0/30 Y71G12B.18/+ | ||
| + lin-65 + + + lin-17/Y71G12B.17 + Y71G12B.18 Y71G12B.27 M01D7.2 +; lin-15A(n767) | Lin-17 | 17/23 M01D7.2/+ | |
| Lin-17 | 18/23 Y71G12B.27/+ | ||
| Lin-17 | 21/23 Y71G12B.18/+ | ||
| Lin-17 | 23/23 Y71G12B.17/+ | ||
| lin(n3542) | + + lin(n3542) lin-15A(n767)/unc-10 dpy-6 + lin-15A(n767) | Unc | 8/8 lin(n3542)/+ |
| + lin(n3542) + lin-15A(n767)/dpy-6 + unc-9 lin-15A(n767) | Unc | 4/40 lin(n3542)/+ | |
| lin(n3628) | lin(n3628) + +/+ dpy-5 unc-13; lin-15A(n767) | Dpy | 0/6 lin(n3628)/+ |
| Unc | 6/6 lin(n3628)/+ | ||
| + lin(n3628) +/unc-11 + dpy-5; lin-15A(n767) | Unc | 1/11 lin(n3628)/+ | |
| Dpy | 5/11 lin(n3628)/+ | ||
| unc-11 + + lin(n3628)/+ unc-73 lin-44 +; lin-15A(n767) | Muv | 3/9 unc-73 lin-44/++ | |
| + + lin(n3628) dpy-5/unc-73 lin-44 + +; lin-15A(n767) | Muv | 0/21 unc-73 lin-44/++ | |
| lin(n3628) + dpy-5/+ unc-38 +; lin-15A(n767) | Muv | 3/7 unc-38/+ | |
| unc-11 lin(n3628) +/+ + unc-38; lin-15A(n767) | Muv | 0/9 unc-38/+ | |
| mep-1 | + mep-1 +/unc-5 + dpy-20; lin-15A(n767) | Unc | 56/57 mep-1/+ |
| Dpy | 2/61 mep-1/+ | ||
| mep-1 + +/+ dpy-20 unc-30; lin-15A(n767) | Dpy | 0/51 mep-1/+ | |
| Unc | 58/58 mep-1/+ | ||
| + + mep-1 +/unc-24 mec-3 + dpy-20; lin-15A(n767) | Unc Mec | 10/12 mep-1/+ | |
| Unc | 17/17 mep-1/+ | ||
| Mec Dpy | 0/8 mep-1/+ | ||
| Dpy | 2/8 mep-1/+ | ||
| + mep-1 dpy-20 +/lin-3 + + unc-22; lin-15A(n767) | Dpy | 5/5 lin-3/+ | |
| Vul | 3/10 mep-1/+ | ||
| + + mep-1+/mec-3 sem-3 + dpy-20; lin-15A(n767) | Mec | 17/17 mep-1/+ | |
| Dpy | 6/13 mep-1/+ | ||
| mys-1 | + mys-1 +/unc-46 + dpy-11; lin-15A | Unc | 3/7 mys-1/+ |
| Dpy | 7/11 mys-1/+ | ||
| trr-1 | + rol-6 + trr-1/dpy-10 + unc-4 +; lin-15A(n767) | Rol | 3/14 unc-4/+ |
| Dpy | 3/3 trr-1/+ | ||
| Unc | 0/8 trr-1/+ | ||
| + trr-1 +/dpy-10 + rol-1; lin-15A(n767) | Rol | 9/20 trr-1/+ | |
| + + trr-1/dpy-10 unc-53 +; lin-15A(n767) | Unc | 0/17 trr-1/+ | |
| + trr-1 +/unc-53 + rol-1; lin-15A(n767) | Unc | 7/10 trr-1/+ | |
| Rol | 7/10 trr-1/+ | ||
| + trr-1 + rol-1/unc-4 + mex-1 +; lin-15A(n767) | Rol | 12/14 mex-1/+ |
Three- and four-factor crosses were performed using standard methods (Brenner 1974). We mapped lin-65 using the Y73E7.2, Y71G12B.2, Y71G12B.17, Y71G12B.18, Y71G12B.27, M01D7.2 DNA sequence polymorphisms present in the CB4856 strain.
Three- and four-factor crosses
Gene . | Genotype of heterozygote . | Phenotype of selected recombinants . | Genotype of selected recombinants (with respect to unselected markers) . |
|---|---|---|---|
| lin-65 | + lin-65 +/bli-3 + lin-17; lin-15A(n767) | Lin-17 | 9/19 lin-65/+ |
| bli-3 + lin-65/+ spe-15 +; lin-15A(n767) | Muv | 10/18 spe-15/+ | |
| + lin-65 lin-17/spe-15 + +; lin-15A(n767) | Lin-17 | 11/11 spe-15/+ | |
| bli-3 + + + lin-65 +/+ Y73E7.2 Y71G12B.2 Y71G12B.17 + Y71G12B.18; lin-15A(n767) | Muv | 4/30 Y73E7.2/+ | |
| Muv | 2/30 Y71G12B.2/+ | ||
| Muv | 1/30 Y71G12B.17/+ | ||
| Muv | 0/30 Y71G12B.18/+ | ||
| + lin-65 + + + lin-17/Y71G12B.17 + Y71G12B.18 Y71G12B.27 M01D7.2 +; lin-15A(n767) | Lin-17 | 17/23 M01D7.2/+ | |
| Lin-17 | 18/23 Y71G12B.27/+ | ||
| Lin-17 | 21/23 Y71G12B.18/+ | ||
| Lin-17 | 23/23 Y71G12B.17/+ | ||
| lin(n3542) | + + lin(n3542) lin-15A(n767)/unc-10 dpy-6 + lin-15A(n767) | Unc | 8/8 lin(n3542)/+ |
| + lin(n3542) + lin-15A(n767)/dpy-6 + unc-9 lin-15A(n767) | Unc | 4/40 lin(n3542)/+ | |
| lin(n3628) | lin(n3628) + +/+ dpy-5 unc-13; lin-15A(n767) | Dpy | 0/6 lin(n3628)/+ |
| Unc | 6/6 lin(n3628)/+ | ||
| + lin(n3628) +/unc-11 + dpy-5; lin-15A(n767) | Unc | 1/11 lin(n3628)/+ | |
| Dpy | 5/11 lin(n3628)/+ | ||
| unc-11 + + lin(n3628)/+ unc-73 lin-44 +; lin-15A(n767) | Muv | 3/9 unc-73 lin-44/++ | |
| + + lin(n3628) dpy-5/unc-73 lin-44 + +; lin-15A(n767) | Muv | 0/21 unc-73 lin-44/++ | |
| lin(n3628) + dpy-5/+ unc-38 +; lin-15A(n767) | Muv | 3/7 unc-38/+ | |
| unc-11 lin(n3628) +/+ + unc-38; lin-15A(n767) | Muv | 0/9 unc-38/+ | |
| mep-1 | + mep-1 +/unc-5 + dpy-20; lin-15A(n767) | Unc | 56/57 mep-1/+ |
| Dpy | 2/61 mep-1/+ | ||
| mep-1 + +/+ dpy-20 unc-30; lin-15A(n767) | Dpy | 0/51 mep-1/+ | |
| Unc | 58/58 mep-1/+ | ||
| + + mep-1 +/unc-24 mec-3 + dpy-20; lin-15A(n767) | Unc Mec | 10/12 mep-1/+ | |
| Unc | 17/17 mep-1/+ | ||
| Mec Dpy | 0/8 mep-1/+ | ||
| Dpy | 2/8 mep-1/+ | ||
| + mep-1 dpy-20 +/lin-3 + + unc-22; lin-15A(n767) | Dpy | 5/5 lin-3/+ | |
| Vul | 3/10 mep-1/+ | ||
| + + mep-1+/mec-3 sem-3 + dpy-20; lin-15A(n767) | Mec | 17/17 mep-1/+ | |
| Dpy | 6/13 mep-1/+ | ||
| mys-1 | + mys-1 +/unc-46 + dpy-11; lin-15A | Unc | 3/7 mys-1/+ |
| Dpy | 7/11 mys-1/+ | ||
| trr-1 | + rol-6 + trr-1/dpy-10 + unc-4 +; lin-15A(n767) | Rol | 3/14 unc-4/+ |
| Dpy | 3/3 trr-1/+ | ||
| Unc | 0/8 trr-1/+ | ||
| + trr-1 +/dpy-10 + rol-1; lin-15A(n767) | Rol | 9/20 trr-1/+ | |
| + + trr-1/dpy-10 unc-53 +; lin-15A(n767) | Unc | 0/17 trr-1/+ | |
| + trr-1 +/unc-53 + rol-1; lin-15A(n767) | Unc | 7/10 trr-1/+ | |
| Rol | 7/10 trr-1/+ | ||
| + trr-1 + rol-1/unc-4 + mex-1 +; lin-15A(n767) | Rol | 12/14 mex-1/+ |
Gene . | Genotype of heterozygote . | Phenotype of selected recombinants . | Genotype of selected recombinants (with respect to unselected markers) . |
|---|---|---|---|
| lin-65 | + lin-65 +/bli-3 + lin-17; lin-15A(n767) | Lin-17 | 9/19 lin-65/+ |
| bli-3 + lin-65/+ spe-15 +; lin-15A(n767) | Muv | 10/18 spe-15/+ | |
| + lin-65 lin-17/spe-15 + +; lin-15A(n767) | Lin-17 | 11/11 spe-15/+ | |
| bli-3 + + + lin-65 +/+ Y73E7.2 Y71G12B.2 Y71G12B.17 + Y71G12B.18; lin-15A(n767) | Muv | 4/30 Y73E7.2/+ | |
| Muv | 2/30 Y71G12B.2/+ | ||
| Muv | 1/30 Y71G12B.17/+ | ||
| Muv | 0/30 Y71G12B.18/+ | ||
| + lin-65 + + + lin-17/Y71G12B.17 + Y71G12B.18 Y71G12B.27 M01D7.2 +; lin-15A(n767) | Lin-17 | 17/23 M01D7.2/+ | |
| Lin-17 | 18/23 Y71G12B.27/+ | ||
| Lin-17 | 21/23 Y71G12B.18/+ | ||
| Lin-17 | 23/23 Y71G12B.17/+ | ||
| lin(n3542) | + + lin(n3542) lin-15A(n767)/unc-10 dpy-6 + lin-15A(n767) | Unc | 8/8 lin(n3542)/+ |
| + lin(n3542) + lin-15A(n767)/dpy-6 + unc-9 lin-15A(n767) | Unc | 4/40 lin(n3542)/+ | |
| lin(n3628) | lin(n3628) + +/+ dpy-5 unc-13; lin-15A(n767) | Dpy | 0/6 lin(n3628)/+ |
| Unc | 6/6 lin(n3628)/+ | ||
| + lin(n3628) +/unc-11 + dpy-5; lin-15A(n767) | Unc | 1/11 lin(n3628)/+ | |
| Dpy | 5/11 lin(n3628)/+ | ||
| unc-11 + + lin(n3628)/+ unc-73 lin-44 +; lin-15A(n767) | Muv | 3/9 unc-73 lin-44/++ | |
| + + lin(n3628) dpy-5/unc-73 lin-44 + +; lin-15A(n767) | Muv | 0/21 unc-73 lin-44/++ | |
| lin(n3628) + dpy-5/+ unc-38 +; lin-15A(n767) | Muv | 3/7 unc-38/+ | |
| unc-11 lin(n3628) +/+ + unc-38; lin-15A(n767) | Muv | 0/9 unc-38/+ | |
| mep-1 | + mep-1 +/unc-5 + dpy-20; lin-15A(n767) | Unc | 56/57 mep-1/+ |
| Dpy | 2/61 mep-1/+ | ||
| mep-1 + +/+ dpy-20 unc-30; lin-15A(n767) | Dpy | 0/51 mep-1/+ | |
| Unc | 58/58 mep-1/+ | ||
| + + mep-1 +/unc-24 mec-3 + dpy-20; lin-15A(n767) | Unc Mec | 10/12 mep-1/+ | |
| Unc | 17/17 mep-1/+ | ||
| Mec Dpy | 0/8 mep-1/+ | ||
| Dpy | 2/8 mep-1/+ | ||
| + mep-1 dpy-20 +/lin-3 + + unc-22; lin-15A(n767) | Dpy | 5/5 lin-3/+ | |
| Vul | 3/10 mep-1/+ | ||
| + + mep-1+/mec-3 sem-3 + dpy-20; lin-15A(n767) | Mec | 17/17 mep-1/+ | |
| Dpy | 6/13 mep-1/+ | ||
| mys-1 | + mys-1 +/unc-46 + dpy-11; lin-15A | Unc | 3/7 mys-1/+ |
| Dpy | 7/11 mys-1/+ | ||
| trr-1 | + rol-6 + trr-1/dpy-10 + unc-4 +; lin-15A(n767) | Rol | 3/14 unc-4/+ |
| Dpy | 3/3 trr-1/+ | ||
| Unc | 0/8 trr-1/+ | ||
| + trr-1 +/dpy-10 + rol-1; lin-15A(n767) | Rol | 9/20 trr-1/+ | |
| + + trr-1/dpy-10 unc-53 +; lin-15A(n767) | Unc | 0/17 trr-1/+ | |
| + trr-1 +/unc-53 + rol-1; lin-15A(n767) | Unc | 7/10 trr-1/+ | |
| Rol | 7/10 trr-1/+ | ||
| + trr-1 + rol-1/unc-4 + mex-1 +; lin-15A(n767) | Rol | 12/14 mex-1/+ |
Three- and four-factor crosses were performed using standard methods (Brenner 1974). We mapped lin-65 using the Y73E7.2, Y71G12B.2, Y71G12B.17, Y71G12B.18, Y71G12B.27, M01D7.2 DNA sequence polymorphisms present in the CB4856 strain.
) and deficiency heterozygotes (Table 3
Deficiency heterozygote mapping
Gene . | Genotype of heterozygote . | Phenotype of heterozygote . |
|---|---|---|
| mep-1 | mep-1/sDf63 unc-31; lin-15A(n767)/+ | Pvl Ste |
| mep-1/sDf62 unc-31; lin-15A(n767)/+ | Pvl Ste | |
| mep-1/sDf10; lin-15A(n767)/+ | WT | |
| trr-1 | rol-6 trr-1/mnDf57; lin-15A(n767) | WT |
| rol-6 trr-1/unc-4 mnDf90; lin-15A(n767) | WT | |
| rol-6 trr-1/mnDf29; lin-15A(n767) | WT | |
| trr-1/unc-4 mnDf87; lin-15A(n767) | Muv |
Gene . | Genotype of heterozygote . | Phenotype of heterozygote . |
|---|---|---|
| mep-1 | mep-1/sDf63 unc-31; lin-15A(n767)/+ | Pvl Ste |
| mep-1/sDf62 unc-31; lin-15A(n767)/+ | Pvl Ste | |
| mep-1/sDf10; lin-15A(n767)/+ | WT | |
| trr-1 | rol-6 trr-1/mnDf57; lin-15A(n767) | WT |
| rol-6 trr-1/unc-4 mnDf90; lin-15A(n767) | WT | |
| rol-6 trr-1/mnDf29; lin-15A(n767) | WT | |
| trr-1/unc-4 mnDf87; lin-15A(n767) | Muv |
Deficiency heterozygotes were constructed as described in materials and methods. WT, wild type; Pvl, protruding vulva; Ste, sterile.
Deficiency heterozygote mapping
Gene . | Genotype of heterozygote . | Phenotype of heterozygote . |
|---|---|---|
| mep-1 | mep-1/sDf63 unc-31; lin-15A(n767)/+ | Pvl Ste |
| mep-1/sDf62 unc-31; lin-15A(n767)/+ | Pvl Ste | |
| mep-1/sDf10; lin-15A(n767)/+ | WT | |
| trr-1 | rol-6 trr-1/mnDf57; lin-15A(n767) | WT |
| rol-6 trr-1/unc-4 mnDf90; lin-15A(n767) | WT | |
| rol-6 trr-1/mnDf29; lin-15A(n767) | WT | |
| trr-1/unc-4 mnDf87; lin-15A(n767) | Muv |
Gene . | Genotype of heterozygote . | Phenotype of heterozygote . |
|---|---|---|
| mep-1 | mep-1/sDf63 unc-31; lin-15A(n767)/+ | Pvl Ste |
| mep-1/sDf62 unc-31; lin-15A(n767)/+ | Pvl Ste | |
| mep-1/sDf10; lin-15A(n767)/+ | WT | |
| trr-1 | rol-6 trr-1/mnDf57; lin-15A(n767) | WT |
| rol-6 trr-1/unc-4 mnDf90; lin-15A(n767) | WT | |
| rol-6 trr-1/mnDf29; lin-15A(n767) | WT | |
| trr-1/unc-4 mnDf87; lin-15A(n767) | Muv |
Deficiency heterozygotes were constructed as described in materials and methods. WT, wild type; Pvl, protruding vulva; Ste, sterile.
) to map these new synMuv genes on their respective linkage groups. While our studies were in progress, mep-1 and lin-65 were independently identified and reported to have a loss-of-function synMuv phenotype (Unhavaithaya et al. 2002; Poulin et al. 2005). Our detailed characterization of the class C synMuv genes mys-1 and trr-1 is presented elsewhere (Ceol and Horvitz 2004). We separated lin-65, lin(n3628), and mep-1 mutations from the parental lin-15A(n767) mutation and found that these mutations alone do not cause extra vulval cells to be produced (Table 4
Interactions of new mutations with class A and class B synMuv mutations
. | . | . | . | Double mutant with class A . | Double mutant with class B . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Single mutant . | lin-15A(n767) . | lin-38 (n751) . | lin-15B(n744) . | lin-35(n745) . | |||||
. | . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . |
| New mutation | lin-65(n3441) | 0 (35) | 3.0 (±0) | 100 (36) | 5.9 (±0.04) | 97 (37) | 5.3 (±0.13) | 4.3 (23) | 3.02 (±0.02) | ND | ND |
| lin(n3628) | 0 (37) | 3.0 (±0) | 71 (41) | 3.9 (±0.14) | 92 (24) | 4.4 (±0.15) | 2.7 (37) | 3.01 (±0.01) | 0 (31) | 3.0 (±0) | |
| mep-1(n3703) | 2.5 (40) | 3.01 (±0.01) | 100 (29) | 6.0 (±0.19) | 100 (36) | 5.9 (±0.29) | 0 (21) | 3.0 (±0) | 0 (25) | 3.0 (±0) | |
| ark-1(n3701) | 0 (33) | 3.0 (±0) | 77 (30) | 4.5 (±0.20) | 56 (34) | 3.8 (±0.14) | 7.8 (26) | 3.06 (±0.04) | 7.4 (27) | 3.07 (±0.05) | |
| gap-1(ga133) | 3.1 (32) | 3.02 (±0.02) | 58 (38) | 3.6 (±0.11) | 76 (37) | 4.4 (±0.17) | 0 (29) | 3.0 (±0) | 0 (30) | 3.0 (±0) | |
| sli-1(n3538) | 0 (25) | 3.0 (±0) | 93 (28) | 4.6 (±0.16) | 30 (27) | 3.3 (±0.11) | 0 (36) | 3.0 (±0) | 4.5 (22) | 3.02 (±0.02) | |
| Class A | lin-15A(n767) | 0 (24)a | 3.0 (±0)a | —c | —c | ND | ND | —c | —c | ||
| lin-38 (n751) | 0 (27)a | 3.0 (±0)a | 0 (32) | 3.0 (±0) | —c | —c | —c | —c | |||
| Class B | lin-15B(n744) | 0 (20) | 3.0 (±0) | ND | ND | 100 (33) | 6.0 (±0) | —c | —c | ||
| lin-35(n745) | 0 (48) | 3.0 (±0) | 100 (22) | 6.0 (±0) | 100 (27) | 6.0 (±0) | 0 (26) | 3.0 (±0) | |||
| Class C | mys-1(n3681) | 8.3 (36)b | 3.06 (±0.03)b | 100 (26)b | 5.04 (±0.14)b | 91 (45)b | 4.40 (±0.13)b | 46 (37) | 3.38 (±0.08) | ND | ND |
| trr-1(n3712) | 13 (89)a | 3.10 (±0.03)a | 74 (54)a | 4.07 (±0.12)a | 79 (14)a | 4.14 (±0.23)a | 50 (38) | 3.38 (±0.07) | 63 (41) | 3.43 (±0.06) | |
. | . | . | . | Double mutant with class A . | Double mutant with class B . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Single mutant . | lin-15A(n767) . | lin-38 (n751) . | lin-15B(n744) . | lin-35(n745) . | |||||
. | . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . |
| New mutation | lin-65(n3441) | 0 (35) | 3.0 (±0) | 100 (36) | 5.9 (±0.04) | 97 (37) | 5.3 (±0.13) | 4.3 (23) | 3.02 (±0.02) | ND | ND |
| lin(n3628) | 0 (37) | 3.0 (±0) | 71 (41) | 3.9 (±0.14) | 92 (24) | 4.4 (±0.15) | 2.7 (37) | 3.01 (±0.01) | 0 (31) | 3.0 (±0) | |
| mep-1(n3703) | 2.5 (40) | 3.01 (±0.01) | 100 (29) | 6.0 (±0.19) | 100 (36) | 5.9 (±0.29) | 0 (21) | 3.0 (±0) | 0 (25) | 3.0 (±0) | |
| ark-1(n3701) | 0 (33) | 3.0 (±0) | 77 (30) | 4.5 (±0.20) | 56 (34) | 3.8 (±0.14) | 7.8 (26) | 3.06 (±0.04) | 7.4 (27) | 3.07 (±0.05) | |
| gap-1(ga133) | 3.1 (32) | 3.02 (±0.02) | 58 (38) | 3.6 (±0.11) | 76 (37) | 4.4 (±0.17) | 0 (29) | 3.0 (±0) | 0 (30) | 3.0 (±0) | |
| sli-1(n3538) | 0 (25) | 3.0 (±0) | 93 (28) | 4.6 (±0.16) | 30 (27) | 3.3 (±0.11) | 0 (36) | 3.0 (±0) | 4.5 (22) | 3.02 (±0.02) | |
| Class A | lin-15A(n767) | 0 (24)a | 3.0 (±0)a | —c | —c | ND | ND | —c | —c | ||
| lin-38 (n751) | 0 (27)a | 3.0 (±0)a | 0 (32) | 3.0 (±0) | —c | —c | —c | —c | |||
| Class B | lin-15B(n744) | 0 (20) | 3.0 (±0) | ND | ND | 100 (33) | 6.0 (±0) | —c | —c | ||
| lin-35(n745) | 0 (48) | 3.0 (±0) | 100 (22) | 6.0 (±0) | 100 (27) | 6.0 (±0) | 0 (26) | 3.0 (±0) | |||
| Class C | mys-1(n3681) | 8.3 (36)b | 3.06 (±0.03)b | 100 (26)b | 5.04 (±0.14)b | 91 (45)b | 4.40 (±0.13)b | 46 (37) | 3.38 (±0.08) | ND | ND |
| trr-1(n3712) | 13 (89)a | 3.10 (±0.03)a | 74 (54)a | 4.07 (±0.12)a | 79 (14)a | 4.14 (±0.23)a | 50 (38) | 3.38 (±0.07) | 63 (41) | 3.43 (±0.06) | |
New synMuv mutations were separated from lin-15A(n767), and double-mutant strains were constructed as described in materials and methods. Because these mutations cause recessive sterility, mep-1(n3703) and trr-1(n3712) homozygotes were derived from heterozygous parents. mep-1(n3703) homozygotes were recognized as the non-Unc progeny of mep-1(n3703)/nT1 n754 heterozygous parents or the non-Gfp progeny of mep-1(n3703)/nT1[qIs51] heterozygous parents. trr-1(n3712) homozygotes were recognized as the non-Gfp progeny of trr-1(n3712)/mIn1[dpy-10(e128) mIs14] heterozygous parents. The lin(n3628) lin-35 strain was marked with unc-13. ND, not determined.
These data are from Table 1 of Ceol and Horvitz (2004).
These data are from Table 3 of Ceol and Horvitz (2004).
These data are found elsewhere in this table.
Interactions of new mutations with class A and class B synMuv mutations
. | . | . | . | Double mutant with class A . | Double mutant with class B . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Single mutant . | lin-15A(n767) . | lin-38 (n751) . | lin-15B(n744) . | lin-35(n745) . | |||||
. | . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . |
| New mutation | lin-65(n3441) | 0 (35) | 3.0 (±0) | 100 (36) | 5.9 (±0.04) | 97 (37) | 5.3 (±0.13) | 4.3 (23) | 3.02 (±0.02) | ND | ND |
| lin(n3628) | 0 (37) | 3.0 (±0) | 71 (41) | 3.9 (±0.14) | 92 (24) | 4.4 (±0.15) | 2.7 (37) | 3.01 (±0.01) | 0 (31) | 3.0 (±0) | |
| mep-1(n3703) | 2.5 (40) | 3.01 (±0.01) | 100 (29) | 6.0 (±0.19) | 100 (36) | 5.9 (±0.29) | 0 (21) | 3.0 (±0) | 0 (25) | 3.0 (±0) | |
| ark-1(n3701) | 0 (33) | 3.0 (±0) | 77 (30) | 4.5 (±0.20) | 56 (34) | 3.8 (±0.14) | 7.8 (26) | 3.06 (±0.04) | 7.4 (27) | 3.07 (±0.05) | |
| gap-1(ga133) | 3.1 (32) | 3.02 (±0.02) | 58 (38) | 3.6 (±0.11) | 76 (37) | 4.4 (±0.17) | 0 (29) | 3.0 (±0) | 0 (30) | 3.0 (±0) | |
| sli-1(n3538) | 0 (25) | 3.0 (±0) | 93 (28) | 4.6 (±0.16) | 30 (27) | 3.3 (±0.11) | 0 (36) | 3.0 (±0) | 4.5 (22) | 3.02 (±0.02) | |
| Class A | lin-15A(n767) | 0 (24)a | 3.0 (±0)a | —c | —c | ND | ND | —c | —c | ||
| lin-38 (n751) | 0 (27)a | 3.0 (±0)a | 0 (32) | 3.0 (±0) | —c | —c | —c | —c | |||
| Class B | lin-15B(n744) | 0 (20) | 3.0 (±0) | ND | ND | 100 (33) | 6.0 (±0) | —c | —c | ||
| lin-35(n745) | 0 (48) | 3.0 (±0) | 100 (22) | 6.0 (±0) | 100 (27) | 6.0 (±0) | 0 (26) | 3.0 (±0) | |||
| Class C | mys-1(n3681) | 8.3 (36)b | 3.06 (±0.03)b | 100 (26)b | 5.04 (±0.14)b | 91 (45)b | 4.40 (±0.13)b | 46 (37) | 3.38 (±0.08) | ND | ND |
| trr-1(n3712) | 13 (89)a | 3.10 (±0.03)a | 74 (54)a | 4.07 (±0.12)a | 79 (14)a | 4.14 (±0.23)a | 50 (38) | 3.38 (±0.07) | 63 (41) | 3.43 (±0.06) | |
. | . | . | . | Double mutant with class A . | Double mutant with class B . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Single mutant . | lin-15A(n767) . | lin-38 (n751) . | lin-15B(n744) . | lin-35(n745) . | |||||
. | . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . |
| New mutation | lin-65(n3441) | 0 (35) | 3.0 (±0) | 100 (36) | 5.9 (±0.04) | 97 (37) | 5.3 (±0.13) | 4.3 (23) | 3.02 (±0.02) | ND | ND |
| lin(n3628) | 0 (37) | 3.0 (±0) | 71 (41) | 3.9 (±0.14) | 92 (24) | 4.4 (±0.15) | 2.7 (37) | 3.01 (±0.01) | 0 (31) | 3.0 (±0) | |
| mep-1(n3703) | 2.5 (40) | 3.01 (±0.01) | 100 (29) | 6.0 (±0.19) | 100 (36) | 5.9 (±0.29) | 0 (21) | 3.0 (±0) | 0 (25) | 3.0 (±0) | |
| ark-1(n3701) | 0 (33) | 3.0 (±0) | 77 (30) | 4.5 (±0.20) | 56 (34) | 3.8 (±0.14) | 7.8 (26) | 3.06 (±0.04) | 7.4 (27) | 3.07 (±0.05) | |
| gap-1(ga133) | 3.1 (32) | 3.02 (±0.02) | 58 (38) | 3.6 (±0.11) | 76 (37) | 4.4 (±0.17) | 0 (29) | 3.0 (±0) | 0 (30) | 3.0 (±0) | |
| sli-1(n3538) | 0 (25) | 3.0 (±0) | 93 (28) | 4.6 (±0.16) | 30 (27) | 3.3 (±0.11) | 0 (36) | 3.0 (±0) | 4.5 (22) | 3.02 (±0.02) | |
| Class A | lin-15A(n767) | 0 (24)a | 3.0 (±0)a | —c | —c | ND | ND | —c | —c | ||
| lin-38 (n751) | 0 (27)a | 3.0 (±0)a | 0 (32) | 3.0 (±0) | —c | —c | —c | —c | |||
| Class B | lin-15B(n744) | 0 (20) | 3.0 (±0) | ND | ND | 100 (33) | 6.0 (±0) | —c | —c | ||
| lin-35(n745) | 0 (48) | 3.0 (±0) | 100 (22) | 6.0 (±0) | 100 (27) | 6.0 (±0) | 0 (26) | 3.0 (±0) | |||
| Class C | mys-1(n3681) | 8.3 (36)b | 3.06 (±0.03)b | 100 (26)b | 5.04 (±0.14)b | 91 (45)b | 4.40 (±0.13)b | 46 (37) | 3.38 (±0.08) | ND | ND |
| trr-1(n3712) | 13 (89)a | 3.10 (±0.03)a | 74 (54)a | 4.07 (±0.12)a | 79 (14)a | 4.14 (±0.23)a | 50 (38) | 3.38 (±0.07) | 63 (41) | 3.43 (±0.06) | |
New synMuv mutations were separated from lin-15A(n767), and double-mutant strains were constructed as described in materials and methods. Because these mutations cause recessive sterility, mep-1(n3703) and trr-1(n3712) homozygotes were derived from heterozygous parents. mep-1(n3703) homozygotes were recognized as the non-Unc progeny of mep-1(n3703)/nT1 n754 heterozygous parents or the non-Gfp progeny of mep-1(n3703)/nT1[qIs51] heterozygous parents. trr-1(n3712) homozygotes were recognized as the non-Gfp progeny of trr-1(n3712)/mIn1[dpy-10(e128) mIs14] heterozygous parents. The lin(n3628) lin-35 strain was marked with unc-13. ND, not determined.
These data are from Table 1 of Ceol and Horvitz (2004).
These data are from Table 3 of Ceol and Horvitz (2004).
These data are found elsewhere in this table.
). Thus, these mutations synergize with lin-15A(n767) and are synMuv mutations.
Interactions with other synMuv mutations:
Since mutations affecting lin-65, lin(n3628), mep-1, gap-1, sli-1, and ark-1 interact synthetically with a class A synMuv mutation, lin-15A(n767), these genes may be either class B or class C synMuv genes or they may define a new synMuv gene class that shares some but not all properties with class B or class C genes. To distinguish between these possiblities, we built double-mutant strains and measured synthetic interactions with lin-65, lin(n3628), mep-1, gap-1, sli-1, and ark-1 mutations. We used the strongest available mutation for each of these genes in these strain constructions. ga133 rather than gap-1(n3535) was used as the gap-1 mutation, because ga133 is a deletion and is considered a null mutation (Hajnal et al. 1997). For the sake of brevity, gap-1(ga133) is referred to as a “new” synMuv mutation hereafter. We quantified synthetic interactions by directly examining the fates of individual P(3–8).p cells (see materials and methods). In wild-type animals three cells invariably adopt vulval fates, whereas in Muv mutants more than three cells adopt vulval fates.
We first measured synthetic interactions with the class A mutation lin-38(n751) and the class B mutations lin-15B(n744) and lin-35(n745) (Table 4). The new synMuv mutations interacted synthetically not only with lin-15A(n767) but also with lin-38(n751), suggesting a general redundancy with the class A synMuv genes. With lin-15B(n744) and lin-35(n745) the new mutations showed weak to no synthetic interaction.
We also investigated whether the new mutations interacted synthetically with the class C mutation trr-1(n3712) (Table 5
Interactions of new mutations with the class C synMuv mutation trr-1(n3712)
. | . | % animals with P8.p vulval fate . | |
|---|---|---|---|
. | . | trr-1(+) . | trr-1(n3712) . |
| + | 0 (many) | 13 (89)a | |
| New mutation | lin-65(n3441) | 0 (35) | 45 (31) |
| lin(n3628) | 0 (37) | 4.2 (24) | |
| mep-1(n3703) | 2.5 (40) | Letb | |
| ark-1(n3701) | 0 (33) | 13 (24) | |
| gap-1(ga133) | 3.1 (32) | 37 (38) | |
| sli-1(n3538) | 0 (25) | 32 (37) | |
| Class A | lin-15A(n767) | 0 (24) | 28 (54) |
| lin-38(n751) | 0 (27) | 36 (14) | |
| Class B | lin-15B(n744) | 0 (20)a | 50 (38)a |
| lin-35(n745) | 0 (48)a | 64 (41)a | |
. | . | % animals with P8.p vulval fate . | |
|---|---|---|---|
. | . | trr-1(+) . | trr-1(n3712) . |
| + | 0 (many) | 13 (89)a | |
| New mutation | lin-65(n3441) | 0 (35) | 45 (31) |
| lin(n3628) | 0 (37) | 4.2 (24) | |
| mep-1(n3703) | 2.5 (40) | Letb | |
| ark-1(n3701) | 0 (33) | 13 (24) | |
| gap-1(ga133) | 3.1 (32) | 37 (38) | |
| sli-1(n3538) | 0 (25) | 32 (37) | |
| Class A | lin-15A(n767) | 0 (24) | 28 (54) |
| lin-38(n751) | 0 (27) | 36 (14) | |
| Class B | lin-15B(n744) | 0 (20)a | 50 (38)a |
| lin-35(n745) | 0 (48)a | 64 (41)a | |
Double-mutant strains were constructed as described in materials and methods. mep-1(n3703) homozygotes were recognized as the non-Unc progeny of mep-1(n3703)/nT1 n754 heterozygous parents or the sterile progeny of mep-1(n3703)/dpy-20(e1282) unc-30(e191) heterozygous parents. trr-1(n3712) homozygotes were recognized as the non-Gfp progeny of trr-1(n3712)/mIn1[dpy-10(e128) mIs14] heterozygous parents.
These data are from Table 1 of Ceol and Horvitz (2004).
We interpret this synthetic lethality as indicating redundancy between mep-1 and trr-1.
Interactions of new mutations with the class C synMuv mutation trr-1(n3712)
. | . | % animals with P8.p vulval fate . | |
|---|---|---|---|
. | . | trr-1(+) . | trr-1(n3712) . |
| + | 0 (many) | 13 (89)a | |
| New mutation | lin-65(n3441) | 0 (35) | 45 (31) |
| lin(n3628) | 0 (37) | 4.2 (24) | |
| mep-1(n3703) | 2.5 (40) | Letb | |
| ark-1(n3701) | 0 (33) | 13 (24) | |
| gap-1(ga133) | 3.1 (32) | 37 (38) | |
| sli-1(n3538) | 0 (25) | 32 (37) | |
| Class A | lin-15A(n767) | 0 (24) | 28 (54) |
| lin-38(n751) | 0 (27) | 36 (14) | |
| Class B | lin-15B(n744) | 0 (20)a | 50 (38)a |
| lin-35(n745) | 0 (48)a | 64 (41)a | |
. | . | % animals with P8.p vulval fate . | |
|---|---|---|---|
. | . | trr-1(+) . | trr-1(n3712) . |
| + | 0 (many) | 13 (89)a | |
| New mutation | lin-65(n3441) | 0 (35) | 45 (31) |
| lin(n3628) | 0 (37) | 4.2 (24) | |
| mep-1(n3703) | 2.5 (40) | Letb | |
| ark-1(n3701) | 0 (33) | 13 (24) | |
| gap-1(ga133) | 3.1 (32) | 37 (38) | |
| sli-1(n3538) | 0 (25) | 32 (37) | |
| Class A | lin-15A(n767) | 0 (24) | 28 (54) |
| lin-38(n751) | 0 (27) | 36 (14) | |
| Class B | lin-15B(n744) | 0 (20)a | 50 (38)a |
| lin-35(n745) | 0 (48)a | 64 (41)a | |
Double-mutant strains were constructed as described in materials and methods. mep-1(n3703) homozygotes were recognized as the non-Unc progeny of mep-1(n3703)/nT1 n754 heterozygous parents or the sterile progeny of mep-1(n3703)/dpy-20(e1282) unc-30(e191) heterozygous parents. trr-1(n3712) homozygotes were recognized as the non-Gfp progeny of trr-1(n3712)/mIn1[dpy-10(e128) mIs14] heterozygous parents.
These data are from Table 1 of Ceol and Horvitz (2004).
We interpret this synthetic lethality as indicating redundancy between mep-1 and trr-1.
). In trr-1(n3712) single mutants, P8.p adopts a vulval cell fate at a low but detectable penetrance (Ceol and Horvitz 2004). We monitored synthetic interactions with trr-1(n3712) for P(3–8).p but report synthetic effects only for P8.p, as this cell is particularly sensitive to cell-fate transformation. lin-65(n3441), mep-1(n3703), gap-1(ga133), and sli-1(n3538) but not lin(n3628) and ark-1(n3701) showed a strong synthetic interaction with trr-1(n3712). In further tests, ark-1(n3701) but not lin(n3628) interacted synthetically with the class C mutation mys-1(n3681): ark-1(n3701); mys-1(n3681) double mutants had a strong synthetic P8.p vulval fate defect (80%, n = 41) as compared to ark-1(n3701) (0%, n = 33) and mys-1(n3681) (8.3%, n = 36) single mutants, whereas the P8.p vulval-fate defect of lin(n3628); mys-1(n3681) (6.7%, n = 30) double mutants was low, like that of lin(n3628) (0%, n = 37) and mys-1(n3681) single mutants. Why ark-1(n3701) interacted with one class C mutation but not another is unclear. It is possible that the synthetic interaction with ark-1(n3701) is sensitive to maternally provided levels of class C synMuv activity and mys-1(n3681), which can be maintained in homozygous strains, provided less maternal activity than did trr-1(n3712), which because of its recessive sterility requires that homozygotes be generated from heterozygous parents.
Most of the new mutations interacted synthetically with class A and class C but not with class B mutations, which indicates that these new mutations are neither class A nor class C mutations. The synthetic interaction of lin(n3628) with class A but not with class B or class C mutations is unusual and is discussed below.
Suppression of let-23 mutations:
Are lin-65, lin(n3628), mep-1, gap-1, sli-1, and ark-1 class B synMuv genes? Neither in combination with class A mutations (Ferguson et al. 1987; Lu and Horvitz 1998; Thomas and Horvitz 1999; Ceol and Horvitz 2001) nor on their own (Table 6
Suppression of the let-23 vulvaless phenotype
. | . | Ave. no. vulval fates (±SE, n) . | |
|---|---|---|---|
. | . | let-23(+) . | let-23(sy97) . |
| + | 3.0 (many) | 0 (±0, 36) | |
| New mutation | lin-65(n3441) | 3.0 (±0, 35)a | 0 (±0, 30) |
| lin(n3628) | 3.0 (±0, 37)a | Letd | |
| mep-1(n3703) | 3.01 (±0.01, 40)a | Letd | |
| ark-1(n3701) | 3.0 (±0, 33)a | 0.10 (±0.05, 34) | |
| gap-1(ga133) | 3.02 (±0.02, 32)a | 3.0 (±0, 26) | |
| sli-1(n3538) | 3.0 (±0, 25)a | 3.0 (±0, 31) | |
| Class A | lin-15A(n767) | 3.0 (±0, 24)b | 0 (±0, 21) |
| lin-38(n751) | 3.0 (±0, 27)b | ND | |
| Class B | lin-15B(n744) | 3.0 (±0, 20)a | 0.23 (±0.08, 26) |
| lin-35(n745) | 3.0 (±0, 48)a | 0.20 (±0.06, 38) | |
| Class C | mys-1(n3681) | 3.06 (±0.03, 36)c | 1.47 (±0.15, 31) |
| trr-1(n3712) | 3.10 (±0.03, 89)b | 0.28 (±0.07, 46) | |
. | . | Ave. no. vulval fates (±SE, n) . | |
|---|---|---|---|
. | . | let-23(+) . | let-23(sy97) . |
| + | 3.0 (many) | 0 (±0, 36) | |
| New mutation | lin-65(n3441) | 3.0 (±0, 35)a | 0 (±0, 30) |
| lin(n3628) | 3.0 (±0, 37)a | Letd | |
| mep-1(n3703) | 3.01 (±0.01, 40)a | Letd | |
| ark-1(n3701) | 3.0 (±0, 33)a | 0.10 (±0.05, 34) | |
| gap-1(ga133) | 3.02 (±0.02, 32)a | 3.0 (±0, 26) | |
| sli-1(n3538) | 3.0 (±0, 25)a | 3.0 (±0, 31) | |
| Class A | lin-15A(n767) | 3.0 (±0, 24)b | 0 (±0, 21) |
| lin-38(n751) | 3.0 (±0, 27)b | ND | |
| Class B | lin-15B(n744) | 3.0 (±0, 20)a | 0.23 (±0.08, 26) |
| lin-35(n745) | 3.0 (±0, 48)a | 0.20 (±0.06, 38) | |
| Class C | mys-1(n3681) | 3.06 (±0.03, 36)c | 1.47 (±0.15, 31) |
| trr-1(n3712) | 3.10 (±0.03, 89)b | 0.28 (±0.07, 46) | |
mep-1(n3703) homozygotes were recognized as the non-Unc progeny of mep-1(n3703)/nT1 n754 heterozygous parents or the sterile progeny of mep-1(n3703)/dpy-20(e1282) unc-30(e191) heterozygous parents. trr-1(n3712) homozygotes were recognized as the non-Gfp progeny of trr-1(n3712) / mIn1[dpy-10(e128) mIs14] heterozygous parents. let-23(sy97) was marked with unc-4(e120), and let-23(sy97) homozygotes were recognized as the Unc non-Gfp progeny of let-23(sy97) unc-4(e120)/mIn1[dpy-10(e128) mIs14] heterozygous parents. ND, not determined because of linkage of these mutations.
These data are from Table 1.
These data are from Table 1 of Ceol and Horvitz (2004).
These data are from Table 3 of Ceol and Horvitz (2004).
Because of the lethality of these animals, we measured the abilities of lin(n3628) and mep-1(n3703) to suppress the Vul phenotype caused by sy10, a let-23 allele that is weaker than sy97. lin(n3628) and mep-1(n3703) were unable to suppress the Vul phenotype of let-23(sy10): lin(n3628); let-23(sy10) double mutants averaged 0.11 vulval fates (n = 27), let-23(sy10); mep-1(n3703) double mutants averaged 0.06 vulval fates (n = 24), and let-23(sy10) single mutants averaged 0.14 vulval fates (n = 21).
Suppression of the let-23 vulvaless phenotype
. | . | Ave. no. vulval fates (±SE, n) . | |
|---|---|---|---|
. | . | let-23(+) . | let-23(sy97) . |
| + | 3.0 (many) | 0 (±0, 36) | |
| New mutation | lin-65(n3441) | 3.0 (±0, 35)a | 0 (±0, 30) |
| lin(n3628) | 3.0 (±0, 37)a | Letd | |
| mep-1(n3703) | 3.01 (±0.01, 40)a | Letd | |
| ark-1(n3701) | 3.0 (±0, 33)a | 0.10 (±0.05, 34) | |
| gap-1(ga133) | 3.02 (±0.02, 32)a | 3.0 (±0, 26) | |
| sli-1(n3538) | 3.0 (±0, 25)a | 3.0 (±0, 31) | |
| Class A | lin-15A(n767) | 3.0 (±0, 24)b | 0 (±0, 21) |
| lin-38(n751) | 3.0 (±0, 27)b | ND | |
| Class B | lin-15B(n744) | 3.0 (±0, 20)a | 0.23 (±0.08, 26) |
| lin-35(n745) | 3.0 (±0, 48)a | 0.20 (±0.06, 38) | |
| Class C | mys-1(n3681) | 3.06 (±0.03, 36)c | 1.47 (±0.15, 31) |
| trr-1(n3712) | 3.10 (±0.03, 89)b | 0.28 (±0.07, 46) | |
. | . | Ave. no. vulval fates (±SE, n) . | |
|---|---|---|---|
. | . | let-23(+) . | let-23(sy97) . |
| + | 3.0 (many) | 0 (±0, 36) | |
| New mutation | lin-65(n3441) | 3.0 (±0, 35)a | 0 (±0, 30) |
| lin(n3628) | 3.0 (±0, 37)a | Letd | |
| mep-1(n3703) | 3.01 (±0.01, 40)a | Letd | |
| ark-1(n3701) | 3.0 (±0, 33)a | 0.10 (±0.05, 34) | |
| gap-1(ga133) | 3.02 (±0.02, 32)a | 3.0 (±0, 26) | |
| sli-1(n3538) | 3.0 (±0, 25)a | 3.0 (±0, 31) | |
| Class A | lin-15A(n767) | 3.0 (±0, 24)b | 0 (±0, 21) |
| lin-38(n751) | 3.0 (±0, 27)b | ND | |
| Class B | lin-15B(n744) | 3.0 (±0, 20)a | 0.23 (±0.08, 26) |
| lin-35(n745) | 3.0 (±0, 48)a | 0.20 (±0.06, 38) | |
| Class C | mys-1(n3681) | 3.06 (±0.03, 36)c | 1.47 (±0.15, 31) |
| trr-1(n3712) | 3.10 (±0.03, 89)b | 0.28 (±0.07, 46) | |
mep-1(n3703) homozygotes were recognized as the non-Unc progeny of mep-1(n3703)/nT1 n754 heterozygous parents or the sterile progeny of mep-1(n3703)/dpy-20(e1282) unc-30(e191) heterozygous parents. trr-1(n3712) homozygotes were recognized as the non-Gfp progeny of trr-1(n3712) / mIn1[dpy-10(e128) mIs14] heterozygous parents. let-23(sy97) was marked with unc-4(e120), and let-23(sy97) homozygotes were recognized as the Unc non-Gfp progeny of let-23(sy97) unc-4(e120)/mIn1[dpy-10(e128) mIs14] heterozygous parents. ND, not determined because of linkage of these mutations.
These data are from Table 1.
These data are from Table 1 of Ceol and Horvitz (2004).
These data are from Table 3 of Ceol and Horvitz (2004).
Because of the lethality of these animals, we measured the abilities of lin(n3628) and mep-1(n3703) to suppress the Vul phenotype caused by sy10, a let-23 allele that is weaker than sy97. lin(n3628) and mep-1(n3703) were unable to suppress the Vul phenotype of let-23(sy10): lin(n3628); let-23(sy10) double mutants averaged 0.11 vulval fates (n = 27), let-23(sy10); mep-1(n3703) double mutants averaged 0.06 vulval fates (n = 24), and let-23(sy10) single mutants averaged 0.14 vulval fates (n = 21).
) do class B mutations suppress the Vul phenotype caused by strong loss-of-function let-23 receptor tyrosine kinase mutations. However, previous studies showed that gap-1 or sli-1 mutations alone can suppress the let-23 Vul phenotype (Jongeward et al. 1995; Hajnal et al. 1997). Together these findings distinguish gap-1 and sli-1 from class B synMuv genes and indicate that let-23 suppression may be used as a criterion in classifying synMuv mutations. We found that mutations affecting lin-65, lin(n3628), mep-1, and ark-1 did not suppress the let-23 Vul phenotype (Table 6), suggesting that these genes are not in the same class as gap-1 and sli-1.
Interactions with ark-1, gap-1, and sli-1 mutations:
gap-1 and sli-1 mutations interact synthetically to produce extra vulval cells (Table 7
Interactions of new mutations with gap-1 and sli-1 mutations
. | . | Double mutant with gap-1(ga133) . | Double mutant with sli-1(n3538) . | ||
|---|---|---|---|---|---|
. | . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . |
| New mutation | lin-65(n3441) | 0 (31) | 3.0 (±0) | 0 (34) | 3.0 (±0) |
| lin(n3628) | 6.1 (33) | 3.05 (±0.03) | 0 (36) | 3.0 (±0) | |
| mep-1(n3703) | 8.3 (36) | 3.07 (±0.04) | 5.6 (36) | 3.03 (±0.02) | |
| ark-1(n3701) | 83 (40) | 4.18 (±0.14) | 48 (29) | 3.48 (±0.11) | |
| gap-1(ga133) | 46 (35) | 3.51 (±0.11) | |||
. | . | Double mutant with gap-1(ga133) . | Double mutant with sli-1(n3538) . | ||
|---|---|---|---|---|---|
. | . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . |
| New mutation | lin-65(n3441) | 0 (31) | 3.0 (±0) | 0 (34) | 3.0 (±0) |
| lin(n3628) | 6.1 (33) | 3.05 (±0.03) | 0 (36) | 3.0 (±0) | |
| mep-1(n3703) | 8.3 (36) | 3.07 (±0.04) | 5.6 (36) | 3.03 (±0.02) | |
| ark-1(n3701) | 83 (40) | 4.18 (±0.14) | 48 (29) | 3.48 (±0.11) | |
| gap-1(ga133) | 46 (35) | 3.51 (±0.11) | |||
Double-mutant strains were constructed as described in materials and methods. mep-1(n3703) homozygotes were recognized as the sterile progeny of mep-1(n3703)/dpy-20(e1282) unc-30(e191) heterozygous parents. trr-1(n3712) homozygotes were recognized as the non-Gfp progeny of trr-1(n3712)/mIn1[dpy-10(e128) mIs14] heterozygous parents.
Interactions of new mutations with gap-1 and sli-1 mutations
. | . | Double mutant with gap-1(ga133) . | Double mutant with sli-1(n3538) . | ||
|---|---|---|---|---|---|
. | . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . |
| New mutation | lin-65(n3441) | 0 (31) | 3.0 (±0) | 0 (34) | 3.0 (±0) |
| lin(n3628) | 6.1 (33) | 3.05 (±0.03) | 0 (36) | 3.0 (±0) | |
| mep-1(n3703) | 8.3 (36) | 3.07 (±0.04) | 5.6 (36) | 3.03 (±0.02) | |
| ark-1(n3701) | 83 (40) | 4.18 (±0.14) | 48 (29) | 3.48 (±0.11) | |
| gap-1(ga133) | 46 (35) | 3.51 (±0.11) | |||
. | . | Double mutant with gap-1(ga133) . | Double mutant with sli-1(n3538) . | ||
|---|---|---|---|---|---|
. | . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . | % >3 vulval fates (n) . | Ave. no. vulval fates (±SE) . |
| New mutation | lin-65(n3441) | 0 (31) | 3.0 (±0) | 0 (34) | 3.0 (±0) |
| lin(n3628) | 6.1 (33) | 3.05 (±0.03) | 0 (36) | 3.0 (±0) | |
| mep-1(n3703) | 8.3 (36) | 3.07 (±0.04) | 5.6 (36) | 3.03 (±0.02) | |
| ark-1(n3701) | 83 (40) | 4.18 (±0.14) | 48 (29) | 3.48 (±0.11) | |
| gap-1(ga133) | 46 (35) | 3.51 (±0.11) | |||
Double-mutant strains were constructed as described in materials and methods. mep-1(n3703) homozygotes were recognized as the sterile progeny of mep-1(n3703)/dpy-20(e1282) unc-30(e191) heterozygous parents. trr-1(n3712) homozygotes were recognized as the non-Gfp progeny of trr-1(n3712)/mIn1[dpy-10(e128) mIs14] heterozygous parents.
). Furthermore, an ark-1 mutation interacts synthetically with these gap-1 and sli-1 mutations, suggesting that all three genes act in parallel in regulating vulval cell fates. Similar synergism of an ark-1 mutation with gap-1 and sli-1 mutations was observed previously (Hopper et al. 2000). By contrast, we observed that the class B synMuv mutations lin-15B(n744) and lin-35(n745) did not interact synthetically with gap-1 or sli-1 mutations (Table 4). This lack of synergism is likely not the result of using weak alleles, as the lin-15B(n744), lin-35(n745), gap-1(ga133), and sli-1(n3538) mutations used in these studies are strong loss-of-function, and possibly null, mutations of their corresponding genes. These results distinguish ark-1 from the class B genes lin-15B and lin-35 Rb and suggest that these class B genes do not act with ark-1 in antagonizing Ras pathway activity. lin-65, lin(n3628), and mep-1 mutations also did not interact synthetically with gap-1(ga133) or sli-1(n3538) (Table 7), revealing a further similarity between lin-65, lin(n3628), and mep-1 mutations and lin-15B and lin-35 Rb class B synMuv mutations.
Molecular identification of lin-65:
We mapped the synMuv gene lin-65 to a small interval between the C. elegans strain CB4856 polymorphisms Y71G12B.17 and Y71G12B.18 (Figure 1A
Molecular cloning of lin-65. (A) The genetic map location of lin-65 on linkage group I (top) and the physical map interval between the C. elegans strain CB4856 polymorphisms Y71G12B.17 and Y71G12B.18 and including lin-65 (bottom). (B) lin-65 gene structure as derived from cDNA and genomic sequences. Shaded boxes indicate coding sequence and an open box indicates the 3′-untranslated region (lin-65 transcripts also contain a 5′-untranslated region that is too small to be viewed in this representation). Predicted translation initiation and termination codons and the poly(A) tail are shown. Arrows above indicate the positions of the lin-65(n3441), lin-65(n3541), and lin-65(n3543) mutations. The fourth exon of the cDNA yk1279h11 is smaller than that of the other five lin-65 cDNAs (the end of the yk1279h11 fourth exon is indicated by a dashed line). The use of an alternative splice donor may have created this shorter fourth exon. However, if the end of the yk1279h11 fourth exon were the site of alternative splicing, a CA and not the typical GT splice donor would have been used. In addition, the end of the yk1279h11-specific fourth exon and the beginning of the fifth exon encode multiple glutamine residues and are highly similar in DNA sequence (see Figure 2). The intervening sequence between two regions of highly similar sequence can be lost because of recombination in bacteria (Robinett et al. 1996). For these reasons we speculate that the apparent alternative splice site at the end of the fourth exon in yk1279h11 may be artifactual and have resulted from an error during the generation or maintenance of this cDNA clone. In support of this possibility: (1) We failed to amplify a shorter-than-wild-type yk1279h11 product in a high-stringency RT–PCR using oligonucleotide primers that flank the putative alternative splice junction, and (2) we failed to amplify any RT–PCR products when an oligonucleotide spanning the putative yk1279h11 alternative splice junction was used in a PCR.
). This interval contains four complete predicted genes, one of which is a micro RNA gene, and portions of three other genes, two of which overlap (C. elegans Sequencing Consortium 1998). We performed RNA-mediated interference (RNAi) to determine if inactivation of any of the three complete, protein-encoding genes would result in a synMuv phenotype. RNAi of Y71G12B.9 caused a Muv phenotype in a lin-15A(n767) but not in a wild-type or lin-15B(n744) background (data not shown). Poulin et al. (2005) independently found that RNAi of Y71G12B.9 caused a synMuv phenotype. We obtained six cDNAs (kindly provided by Yuji Kohara and co-workers) and compared the sequences of these clones with genomic sequence to determine a gene structure for Y71G12B.9 (Figure 1B). One clone had an SL1 and two had an SL2 splice-leader sequence. The presence of an SL2 splice leader suggests that Y71G12B.9 is a downstream gene in an operon (Zorio et al. 1994). The predicted initiator methionine codon of the SL2-spliced Y71G12B.9 cDNAs lies just downstream of the trans-splice site. The open reading frame beginning with this initiator methionine encodes a 728-amino-acid protein (Figure 2
lin-65 cDNA sequence indicating differences among individual lin-65 cDNAs. SL1 and SL2 splice-leader sequences are italicized. The SL1 leader, as observed with one cDNA, is spliced two nucleotides downstream of the site at which the SL2 leader is spliced, as observed with two independently derived cDNAs. Intron positions are indicated by carats. The translation termination codon is underlined. Sites of alternative polyadenylation are indicated with solid arrowheads. The predicted LIN-65 protein is shown beneath. The SL2-spliced cDNAs are predicted to encode a 728-amino-acid protein. The SL1-spliced cDNA cannot encode the predicted initiator methionine of the 728-amino-acid protein; it may use the underlined methionine codon to initiate synthesis of a 691-amino-acid protein. The alternatively spliced cDNA yk1279h11 is predicted to encode a protein lacking amino acids 421–481 of the 728-amino-acid protein, although, as described in the Figure 1 legend, the alternative splicing of yk1279h11 is likely to be artifactual. The site at which the putative fourth exon in yk1279h11 ends is indicated with an open arrowhead. This end is juxtaposed to the beginning of the fifth exon to give a CAGCAACAA/CAACAAAAT junction sequence.
). The SL1 trans-splice site is downstream from that of SL2, and the single SL1-spliced cDNA lacks the initiator methionine corresponding to the 728-amino-acid predicted protein. The open reading frames defined by the first three potential initiator methionine codons of the SL1-spliced cDNA are all short (≤16 codons). If the fourth potential initiator methionine codon were used, a 691-amino-acid protein lacking the first 37 amino acids of the 728-amino-acid protein described above would be synthesized. Expression under the control of the C. elegans heat-shock promoters of a cDNA predicted to encode the 728-amino-acid protein rescued the Muv phenotype of lin-65 mutants: two transgenic lines of lin-65(n3441); lin-15A(n767) mutants containing Phs:lin-65 transgenes were 0% (n = 73) and 2.0% (n = 49) non-Muv without heat shock but were 71% (n = 68) and 67% (n = 30) non-Muv, respectively, following heat-shock treatment.
We determined the sequence of Y71G12B.9 in lin-65(n3441), lin-65(n3541), and lin-65(n3543) mutants. lin-65(n3441) and lin-65(n3541) contain identical nonsense mutations predicted to truncate the Y71G12B.9 protein after 533 of the 728 amino acids. It is unlikely that lin-65(n3441) and lin-65(n3541) were caused by the same mutational event, since they were isolated from independently mutagenized and screened P0 animals. lin-65(n3543) contains a missense mutation that changes a polar serine residue to a nonpolar leucine (S720L). The map position, RNAi phenocopy, and cDNA rescue data as well as the mutant allele sequences indicate that Y71G12B.9 is lin-65.
The 728-amino-acid LIN-65 protein is rich in acidic amino acids (Figure 2). Over 7 and 10% of the total number of amino acids are aspartates and glutamates, respectively, and these acidic amino acids are found both in clusters and dispersed throughout LIN-65. BLAST searches (Altschul et al. 1990) with LIN-65 identified proteins from mammalian and other species that are similarly acid rich. Because the similarity between LIN-65 and these proteins is primarily limited to acidic residues and not to specific protein domains (data not shown), it is difficult to predict whether these proteins are functional orthologs of LIN-65. As described above, mutations in lin-65 and lin-35 Rb show similar genetic interactions, suggesting that lin-65 is a class B synMuv gene that acts in the lin-35 Rb pathway. Protein complexes purified from Drosophila extracts and analogous to a class B synMuv complex (M. M. Harrison and H. R. Horvitz, unpublished observations) have not been reported to contain LIN-65-like proteins (Korenjak et al. 2004; Lewis et al. 2004). It is possible that LIN-65 and LIN-65 orthologs act upstream of class B synMuv and analogous complexes to promote complex activity or act downstream as effectors of these complexes.
Sequences of synMuv mutations:
We determined DNA sequences of 41 mutant synMuv genes identified in our screen; 4 of these mutant genes had two distinct mutations (Table 8
Sequences of new mutations of class B and class C synMuv proteins
Protein . | Class . | No. amino acids . | Protein similarities and domainsa . |
|---|---|---|---|
| A. Features of synMuv proteins | |||
| DPL-1 | B | 598 | Similar to DP family transcription factors; contains DNA- and E2F-binding domains. |
| EFL-1 | B | 342 | Similar to E2F family transcription factors; contains DNA-, DP- and Rb-binding domains. |
| LET-418 | B | 1829 | Similar to Mi-2 family ATP-dependent chromatin remodeling enzymes; contains chromodomains, PHD finger motifs, and a helicase domainb |
| LIN-9 | B | LIN-9L: 644 LIN-9S: 642 | Similar to Drosophila Mip130 DNA replication and Aly cell cycle regulators and mammalian proteins of unknown function. |
| LIN-13 | B | 2248 | 24 predicted Zn-finger motifs. |
| LIN-35 | B | 961 | Similar to Retinoblastoma (pRb) family transcriptional regulators; contains “pocket” interaction domain. |
| LIN-36 | B | 962 | THAP domain, C/H-rich and Q-rich regions. |
| LIN-52 | B | 161 | Similar to Drosophila and mammalian proteins of unknown function. |
| LIN-53 | B | 417 | Similar to Drosophila p55, mammalian RbAp48 subunits of chromatin remodeling, and histone deacetylase complexes; contains WD repeats. |
| LIN-65 | B | 728 | Acid rich |
| MEP-1 | B | 853 | Six Zn-finger motifs. |
| MYS-1 | C | 458 | Similar to MYST family histone acetyltransferases; contains chromodomain and acetyltransferase domain. |
| SLI-1 | sli-1 | 582 | Similar to Cbl family ubiquitination-promoting proteins; contains SH2 domain and RING finger motif. |
| TRR-1 | C | 4064c | Similar to mammalian TRRAP transcriptional regulator. |
Protein . | Class . | No. amino acids . | Protein similarities and domainsa . |
|---|---|---|---|
| A. Features of synMuv proteins | |||
| DPL-1 | B | 598 | Similar to DP family transcription factors; contains DNA- and E2F-binding domains. |
| EFL-1 | B | 342 | Similar to E2F family transcription factors; contains DNA-, DP- and Rb-binding domains. |
| LET-418 | B | 1829 | Similar to Mi-2 family ATP-dependent chromatin remodeling enzymes; contains chromodomains, PHD finger motifs, and a helicase domainb |
| LIN-9 | B | LIN-9L: 644 LIN-9S: 642 | Similar to Drosophila Mip130 DNA replication and Aly cell cycle regulators and mammalian proteins of unknown function. |
| LIN-13 | B | 2248 | 24 predicted Zn-finger motifs. |
| LIN-35 | B | 961 | Similar to Retinoblastoma (pRb) family transcriptional regulators; contains “pocket” interaction domain. |
| LIN-36 | B | 962 | THAP domain, C/H-rich and Q-rich regions. |
| LIN-52 | B | 161 | Similar to Drosophila and mammalian proteins of unknown function. |
| LIN-53 | B | 417 | Similar to Drosophila p55, mammalian RbAp48 subunits of chromatin remodeling, and histone deacetylase complexes; contains WD repeats. |
| LIN-65 | B | 728 | Acid rich |
| MEP-1 | B | 853 | Six Zn-finger motifs. |
| MYS-1 | C | 458 | Similar to MYST family histone acetyltransferases; contains chromodomain and acetyltransferase domain. |
| SLI-1 | sli-1 | 582 | Similar to Cbl family ubiquitination-promoting proteins; contains SH2 domain and RING finger motif. |
| TRR-1 | C | 4064c | Similar to mammalian TRRAP transcriptional regulator. |
| Mutation . | Wild-type sequence . | Mutant sequence . | Substitution, splice site change, or aberration . | Domain affected by missense mutation . |
|---|---|---|---|---|
| B. Allele sequences | ||||
| dpl-1(n3643)d | TAT | TAA | Y341ochre | — |
| GGC | CGC | G533R | Unknown | |
| efl-1(n3639)e | CAA | TAA | Q175ochre | — |
| let-418(n3536) | CCT | CTT | P675L | Helicase/ATPase |
| let-418(n3626) | GGT | AGT | G1006S | Helicase/ATPase |
| let-418(n3629) | TCC | TTC | S925F | Helicase/ATPase |
| let-418(n3634) | TGG | TAG | W1128amber | — |
| let-418(n3635) | CAG | TAG | Q1594amber | — |
| let-418(n3636) | ACT | TCT | T807S | Helicase/ATPase |
| TGG | TGA | W1329opal | — | |
| let-418(n3719) | TGG | TAG | W295amber | — |
| lin-9(n3631) | CAA | TAA | LIN-9L: Q594ochre | — |
| LIN-9S: Q592ochre | — | |||
| lin-9(n3675) | GAT | AAT | LIN-9L: D305N | Unknown |
| LIN-9S: D303N | Unknown | |||
| lin-9(n3767) | CAG | TAG | LIN-9L: Q509amber | — |
| LIN-9S: Q507amber | — | |||
| lin-13(n3642) | CAT | TAT | H832Y | Zn finger |
| lin-13(n3673) | CAG | TAG | Q1988amber | — |
| lin-13(n3674) | CGA | TGA | R1250opal | — |
| lin-13(n3726) | GGA | GAA | G229E | Unknown |
| lin-35(n3763)f | GCA | GTA | A555V | |
| TTG AAA AAG | TTG AAA AAA G | K594 frameshift and truncation after 611 a.a. | — | |
| lin-36(n3671) | CAT | CCT | H284P | C/H-rich region |
| GAA | AAA | E424K | Unknown | |
| lin-36(n3672) | CAG | TAG | Q467amber | — |
| lin-36(n3765)g | GCT | GTT | A242V | C/H-rich region |
| lin-52(n3718)h | CAG | TAG | Q31amber | — |
| lin-53(n3448) | AGT | ATT | S384I | WD repeat |
| lin-53(n3521) | GAA | AAA | E174K | WD repeat |
| lin-53(n3622) | AAG/gtatgtgt | AAG/atatgtgt | Exon 1 donor | — |
| lin-53(n3623) | TGG | TAG | W337amber | — |
| lin-65(n3441) | TGG | TGA | W534amber | — |
| lin-65(n3541) | TGG | TGA | W534amber | — |
| lin-65(n3543) | TCG | TTG | S720L | Unknown |
| mep-1(n3680) | AGT | AAT | S309N | Unknown |
| mep-1(n3702) | CAG | TAG | Q706amber | — |
| mep-1(n3703) | CTT/gtaagttt | CTT/ataagttt | Exon 3 donor | — |
| mys-1(n3681)i | GGA | AGA | G341R | Acetyltransferase |
| sli-1(n3538) | TCA | TTA | S305L | SH2 |
| sli-1(n3544) | ttttccag/AAA | ttttccaa/AAA | Exon 6 acceptor | — |
| sli-1(n3683) | ttttttag/GAT | ttttttaa/GAT | Exon 4 acceptor | — |
| trr-1(n3630)j | TGG | TAG | W2064amber | — |
| trr-1(n3637)j | CAG | TAG | Q3444amber | — |
| trr-1(n3704)j | CAA | TAA | Q694ochre | — |
| trr-1(n3708)j | CGA | TGA | R1248opal | — |
| trr-1(n3709)j | CGA | TGA | R2550opal | — |
| trr-1(n3712)j | TGG | TAG | W2505amber | — |
| Mutation . | Wild-type sequence . | Mutant sequence . | Substitution, splice site change, or aberration . | Domain affected by missense mutation . |
|---|---|---|---|---|
| B. Allele sequences | ||||
| dpl-1(n3643)d | TAT | TAA | Y341ochre | — |
| GGC | CGC | G533R | Unknown | |
| efl-1(n3639)e | CAA | TAA | Q175ochre | — |
| let-418(n3536) | CCT | CTT | P675L | Helicase/ATPase |
| let-418(n3626) | GGT | AGT | G1006S | Helicase/ATPase |
| let-418(n3629) | TCC | TTC | S925F | Helicase/ATPase |
| let-418(n3634) | TGG | TAG | W1128amber | — |
| let-418(n3635) | CAG | TAG | Q1594amber | — |
| let-418(n3636) | ACT | TCT | T807S | Helicase/ATPase |
| TGG | TGA | W1329opal | — | |
| let-418(n3719) | TGG | TAG | W295amber | — |
| lin-9(n3631) | CAA | TAA | LIN-9L: Q594ochre | — |
| LIN-9S: Q592ochre | — | |||
| lin-9(n3675) | GAT | AAT | LIN-9L: D305N | Unknown |
| LIN-9S: D303N | Unknown | |||
| lin-9(n3767) | CAG | TAG | LIN-9L: Q509amber | — |
| LIN-9S: Q507amber | — | |||
| lin-13(n3642) | CAT | TAT | H832Y | Zn finger |
| lin-13(n3673) | CAG | TAG | Q1988amber | — |
| lin-13(n3674) | CGA | TGA | R1250opal | — |
| lin-13(n3726) | GGA | GAA | G229E | Unknown |
| lin-35(n3763)f | GCA | GTA | A555V | |
| TTG AAA AAG | TTG AAA AAA G | K594 frameshift and truncation after 611 a.a. | — | |
| lin-36(n3671) | CAT | CCT | H284P | C/H-rich region |
| GAA | AAA | E424K | Unknown | |
| lin-36(n3672) | CAG | TAG | Q467amber | — |
| lin-36(n3765)g | GCT | GTT | A242V | C/H-rich region |
| lin-52(n3718)h | CAG | TAG | Q31amber | — |
| lin-53(n3448) | AGT | ATT | S384I | WD repeat |
| lin-53(n3521) | GAA | AAA | E174K | WD repeat |
| lin-53(n3622) | AAG/gtatgtgt | AAG/atatgtgt | Exon 1 donor | — |
| lin-53(n3623) | TGG | TAG | W337amber | — |
| lin-65(n3441) | TGG | TGA | W534amber | — |
| lin-65(n3541) | TGG | TGA | W534amber | — |
| lin-65(n3543) | TCG | TTG | S720L | Unknown |
| mep-1(n3680) | AGT | AAT | S309N | Unknown |
| mep-1(n3702) | CAG | TAG | Q706amber | — |
| mep-1(n3703) | CTT/gtaagttt | CTT/ataagttt | Exon 3 donor | — |
| mys-1(n3681)i | GGA | AGA | G341R | Acetyltransferase |
| sli-1(n3538) | TCA | TTA | S305L | SH2 |
| sli-1(n3544) | ttttccag/AAA | ttttccaa/AAA | Exon 6 acceptor | — |
| sli-1(n3683) | ttttttag/GAT | ttttttaa/GAT | Exon 4 acceptor | — |
| trr-1(n3630)j | TGG | TAG | W2064amber | — |
| trr-1(n3637)j | CAG | TAG | Q3444amber | — |
| trr-1(n3704)j | CAA | TAA | Q694ochre | — |
| trr-1(n3708)j | CGA | TGA | R1248opal | — |
| trr-1(n3709)j | CGA | TGA | R2550opal | — |
| trr-1(n3712)j | TGG | TAG | W2505amber | — |
The synMuv proteins described are limited to those for which we obtained mutant allele sequence; this is not a comprehensive listing of synMuv proteins. In the “Wild-type sequence” and “Mutant sequence” columns, exon and intron sequences are denoted by uppercase and lowercase letters, respectively. Nucleotides altered by the mutations are underlined.
Molecular descriptions of the proteins listed were obtained from the following sources: DPL-1, and EFL-1, Ceol and Horvitz (2001) and Page et al. (2001); LET-418, Solari and Ahringer (2000) and von Zelewsky et al. (2000); LIN-9, Beitel et al. (2000); LIN-13, Melendez and Greenwald (2000); LIN-35 and LIN-53, Lu and Horvitz (1998); LIN-36, Thomas and Horvitz (1999) and Reddy and Villeneuve (2004); LIN-52, Thomas et al. (2003); MEP-1, Belfiore et al. (2002); SLI-1, Yoon et al. (1995); MYS-1 and TRR-1, Ceol and Horvitz (2004).
The predicted LET-418 protein contains a sequence that is annotated as a helicase domain (see www.wormbase.org). This domain was originally identified in helicases but has since been found in nonhelicase proteins. Many of these proteins share a common ATPase activity, and this domain contains residues that are important for ATP binding and hydrolysis.
Because of alternative splicing, trr-1 encodes proteins that may range in length between 4054 and 4064 amino acids (Ceol and Horvitz 2004).
These data are from Figure 1 of Ceol and Horvitz (2001).
These data are from Figure 4 of Ceol and Horvitz (2001).
The adenosine inserted by the lin-35(n3763) frameshift mutation is not underlined, because it is unclear which adenosine in the adenosine repeat was inserted.
In addition to the missense mutation described, we found an additional mutation associated with lin-36(n3765). This mutation, AG/gtaagaagaaaagc to AG/gtaagaagaaaagt, is present in the third intron of lin-36 and creates a possible splice-donor sequence. If this splice-donor were used, an in-frame ochre (TAA) stop codon would be encountered, truncating the LIN-36 protein after 261 amino acids.
These data are from Figure 3 of Thomas et al. (2003).
These data are from Figure 2 of Ceol and Horvitz (2004).
These data are from Figure 1 of Ceol and Horvitz (2004).
Sequences of new mutations of class B and class C synMuv proteins
Protein . | Class . | No. amino acids . | Protein similarities and domainsa . |
|---|---|---|---|
| A. Features of synMuv proteins | |||
| DPL-1 | B | 598 | Similar to DP family transcription factors; contains DNA- and E2F-binding domains. |
| EFL-1 | B | 342 | Similar to E2F family transcription factors; contains DNA-, DP- and Rb-binding domains. |
| LET-418 | B | 1829 | Similar to Mi-2 family ATP-dependent chromatin remodeling enzymes; contains chromodomains, PHD finger motifs, and a helicase domainb |
| LIN-9 | B | LIN-9L: 644 LIN-9S: 642 | Similar to Drosophila Mip130 DNA replication and Aly cell cycle regulators and mammalian proteins of unknown function. |
| LIN-13 | B | 2248 | 24 predicted Zn-finger motifs. |
| LIN-35 | B | 961 | Similar to Retinoblastoma (pRb) family transcriptional regulators; contains “pocket” interaction domain. |
| LIN-36 | B | 962 | THAP domain, C/H-rich and Q-rich regions. |
| LIN-52 | B | 161 | Similar to Drosophila and mammalian proteins of unknown function. |
| LIN-53 | B | 417 | Similar to Drosophila p55, mammalian RbAp48 subunits of chromatin remodeling, and histone deacetylase complexes; contains WD repeats. |
| LIN-65 | B | 728 | Acid rich |
| MEP-1 | B | 853 | Six Zn-finger motifs. |
| MYS-1 | C | 458 | Similar to MYST family histone acetyltransferases; contains chromodomain and acetyltransferase domain. |
| SLI-1 | sli-1 | 582 | Similar to Cbl family ubiquitination-promoting proteins; contains SH2 domain and RING finger motif. |
| TRR-1 | C | 4064c | Similar to mammalian TRRAP transcriptional regulator. |
Protein . | Class . | No. amino acids . | Protein similarities and domainsa . |
|---|---|---|---|
| A. Features of synMuv proteins | |||
| DPL-1 | B | 598 | Similar to DP family transcription factors; contains DNA- and E2F-binding domains. |
| EFL-1 | B | 342 | Similar to E2F family transcription factors; contains DNA-, DP- and Rb-binding domains. |
| LET-418 | B | 1829 | Similar to Mi-2 family ATP-dependent chromatin remodeling enzymes; contains chromodomains, PHD finger motifs, and a helicase domainb |
| LIN-9 | B | LIN-9L: 644 LIN-9S: 642 | Similar to Drosophila Mip130 DNA replication and Aly cell cycle regulators and mammalian proteins of unknown function. |
| LIN-13 | B | 2248 | 24 predicted Zn-finger motifs. |
| LIN-35 | B | 961 | Similar to Retinoblastoma (pRb) family transcriptional regulators; contains “pocket” interaction domain. |
| LIN-36 | B | 962 | THAP domain, C/H-rich and Q-rich regions. |
| LIN-52 | B | 161 | Similar to Drosophila and mammalian proteins of unknown function. |
| LIN-53 | B | 417 | Similar to Drosophila p55, mammalian RbAp48 subunits of chromatin remodeling, and histone deacetylase complexes; contains WD repeats. |
| LIN-65 | B | 728 | Acid rich |
| MEP-1 | B | 853 | Six Zn-finger motifs. |
| MYS-1 | C | 458 | Similar to MYST family histone acetyltransferases; contains chromodomain and acetyltransferase domain. |
| SLI-1 | sli-1 | 582 | Similar to Cbl family ubiquitination-promoting proteins; contains SH2 domain and RING finger motif. |
| TRR-1 | C | 4064c | Similar to mammalian TRRAP transcriptional regulator. |
| Mutation . | Wild-type sequence . | Mutant sequence . | Substitution, splice site change, or aberration . | Domain affected by missense mutation . |
|---|---|---|---|---|
| B. Allele sequences | ||||
| dpl-1(n3643)d | TAT | TAA | Y341ochre | — |
| GGC | CGC | G533R | Unknown | |
| efl-1(n3639)e | CAA | TAA | Q175ochre | — |
| let-418(n3536) | CCT | CTT | P675L | Helicase/ATPase |
| let-418(n3626) | GGT | AGT | G1006S | Helicase/ATPase |
| let-418(n3629) | TCC | TTC | S925F | Helicase/ATPase |
| let-418(n3634) | TGG | TAG | W1128amber | — |
| let-418(n3635) | CAG | TAG | Q1594amber | — |
| let-418(n3636) | ACT | TCT | T807S | Helicase/ATPase |
| TGG | TGA | W1329opal | — | |
| let-418(n3719) | TGG | TAG | W295amber | — |
| lin-9(n3631) | CAA | TAA | LIN-9L: Q594ochre | — |
| LIN-9S: Q592ochre | — | |||
| lin-9(n3675) | GAT | AAT | LIN-9L: D305N | Unknown |
| LIN-9S: D303N | Unknown | |||
| lin-9(n3767) | CAG | TAG | LIN-9L: Q509amber | — |
| LIN-9S: Q507amber | — | |||
| lin-13(n3642) | CAT | TAT | H832Y | Zn finger |
| lin-13(n3673) | CAG | TAG | Q1988amber | — |
| lin-13(n3674) | CGA | TGA | R1250opal | — |
| lin-13(n3726) | GGA | GAA | G229E | Unknown |
| lin-35(n3763)f | GCA | GTA | A555V | |
| TTG AAA AAG | TTG AAA AAA G | K594 frameshift and truncation after 611 a.a. | — | |
| lin-36(n3671) | CAT | CCT | H284P | C/H-rich region |
| GAA | AAA | E424K | Unknown | |
| lin-36(n3672) | CAG | TAG | Q467amber | — |
| lin-36(n3765)g | GCT | GTT | A242V | C/H-rich region |
| lin-52(n3718)h | CAG | TAG | Q31amber | — |
| lin-53(n3448) | AGT | ATT | S384I | WD repeat |
| lin-53(n3521) | GAA | AAA | E174K | WD repeat |
| lin-53(n3622) | AAG/gtatgtgt | AAG/atatgtgt | Exon 1 donor | — |
| lin-53(n3623) | TGG | TAG | W337amber | — |
| lin-65(n3441) | TGG | TGA | W534amber | — |
| lin-65(n3541) | TGG | TGA | W534amber | — |
| lin-65(n3543) | TCG | TTG | S720L | Unknown |
| mep-1(n3680) | AGT | AAT | S309N | Unknown |
| mep-1(n3702) | CAG | TAG | Q706amber | — |
| mep-1(n3703) | CTT/gtaagttt | CTT/ataagttt | Exon 3 donor | — |
| mys-1(n3681)i | GGA | AGA | G341R | Acetyltransferase |
| sli-1(n3538) | TCA | TTA | S305L | SH2 |
| sli-1(n3544) | ttttccag/AAA | ttttccaa/AAA | Exon 6 acceptor | — |
| sli-1(n3683) | ttttttag/GAT | ttttttaa/GAT | Exon 4 acceptor | — |
| trr-1(n3630)j | TGG | TAG | W2064amber | — |
| trr-1(n3637)j | CAG | TAG | Q3444amber | — |
| trr-1(n3704)j | CAA | TAA | Q694ochre | — |
| trr-1(n3708)j | CGA | TGA | R1248opal | — |
| trr-1(n3709)j | CGA | TGA | R2550opal | — |
| trr-1(n3712)j | TGG | TAG | W2505amber | — |
| Mutation . | Wild-type sequence . | Mutant sequence . | Substitution, splice site change, or aberration . | Domain affected by missense mutation . |
|---|---|---|---|---|
| B. Allele sequences | ||||
| dpl-1(n3643)d | TAT | TAA | Y341ochre | — |
| GGC | CGC | G533R | Unknown | |
| efl-1(n3639)e | CAA | TAA | Q175ochre | — |
| let-418(n3536) | CCT | CTT | P675L | Helicase/ATPase |
| let-418(n3626) | GGT | AGT | G1006S | Helicase/ATPase |
| let-418(n3629) | TCC | TTC | S925F | Helicase/ATPase |
| let-418(n3634) | TGG | TAG | W1128amber | — |
| let-418(n3635) | CAG | TAG | Q1594amber | — |
| let-418(n3636) | ACT | TCT | T807S | Helicase/ATPase |
| TGG | TGA | W1329opal | — | |
| let-418(n3719) | TGG | TAG | W295amber | — |
| lin-9(n3631) | CAA | TAA | LIN-9L: Q594ochre | — |
| LIN-9S: Q592ochre | — | |||
| lin-9(n3675) | GAT | AAT | LIN-9L: D305N | Unknown |
| LIN-9S: D303N | Unknown | |||
| lin-9(n3767) | CAG | TAG | LIN-9L: Q509amber | — |
| LIN-9S: Q507amber | — | |||
| lin-13(n3642) | CAT | TAT | H832Y | Zn finger |
| lin-13(n3673) | CAG | TAG | Q1988amber | — |
| lin-13(n3674) | CGA | TGA | R1250opal | — |
| lin-13(n3726) | GGA | GAA | G229E | Unknown |
| lin-35(n3763)f | GCA | GTA | A555V | |
| TTG AAA AAG | TTG AAA AAA G | K594 frameshift and truncation after 611 a.a. | — | |
| lin-36(n3671) | CAT | CCT | H284P | C/H-rich region |
| GAA | AAA | E424K | Unknown | |
| lin-36(n3672) | CAG | TAG | Q467amber | — |
| lin-36(n3765)g | GCT | GTT | A242V | C/H-rich region |
| lin-52(n3718)h | CAG | TAG | Q31amber | — |
| lin-53(n3448) | AGT | ATT | S384I | WD repeat |
| lin-53(n3521) | GAA | AAA | E174K | WD repeat |
| lin-53(n3622) | AAG/gtatgtgt | AAG/atatgtgt | Exon 1 donor | — |
| lin-53(n3623) | TGG | TAG | W337amber | — |
| lin-65(n3441) | TGG | TGA | W534amber | — |
| lin-65(n3541) | TGG | TGA | W534amber | — |
| lin-65(n3543) | TCG | TTG | S720L | Unknown |
| mep-1(n3680) | AGT | AAT | S309N | Unknown |
| mep-1(n3702) | CAG | TAG | Q706amber | — |
| mep-1(n3703) | CTT/gtaagttt | CTT/ataagttt | Exon 3 donor | — |
| mys-1(n3681)i | GGA | AGA | G341R | Acetyltransferase |
| sli-1(n3538) | TCA | TTA | S305L | SH2 |
| sli-1(n3544) | ttttccag/AAA | ttttccaa/AAA | Exon 6 acceptor | — |
| sli-1(n3683) | ttttttag/GAT | ttttttaa/GAT | Exon 4 acceptor | — |
| trr-1(n3630)j | TGG | TAG | W2064amber | — |
| trr-1(n3637)j | CAG | TAG | Q3444amber | — |
| trr-1(n3704)j | CAA | TAA | Q694ochre | — |
| trr-1(n3708)j | CGA | TGA | R1248opal | — |
| trr-1(n3709)j | CGA | TGA | R2550opal | — |
| trr-1(n3712)j | TGG | TAG | W2505amber | — |
The synMuv proteins described are limited to those for which we obtained mutant allele sequence; this is not a comprehensive listing of synMuv proteins. In the “Wild-type sequence” and “Mutant sequence” columns, exon and intron sequences are denoted by uppercase and lowercase letters, respectively. Nucleotides altered by the mutations are underlined.
Molecular descriptions of the proteins listed were obtained from the following sources: DPL-1, and EFL-1, Ceol and Horvitz (2001) and Page et al. (2001); LET-418, Solari and Ahringer (2000) and von Zelewsky et al. (2000); LIN-9, Beitel et al. (2000); LIN-13, Melendez and Greenwald (2000); LIN-35 and LIN-53, Lu and Horvitz (1998); LIN-36, Thomas and Horvitz (1999) and Reddy and Villeneuve (2004); LIN-52, Thomas et al. (2003); MEP-1, Belfiore et al. (2002); SLI-1, Yoon et al. (1995); MYS-1 and TRR-1, Ceol and Horvitz (2004).
The predicted LET-418 protein contains a sequence that is annotated as a helicase domain (see www.wormbase.org). This domain was originally identified in helicases but has since been found in nonhelicase proteins. Many of these proteins share a common ATPase activity, and this domain contains residues that are important for ATP binding and hydrolysis.
Because of alternative splicing, trr-1 encodes proteins that may range in length between 4054 and 4064 amino acids (Ceol and Horvitz 2004).
These data are from Figure 1 of Ceol and Horvitz (2001).
These data are from Figure 4 of Ceol and Horvitz (2001).
The adenosine inserted by the lin-35(n3763) frameshift mutation is not underlined, because it is unclear which adenosine in the adenosine repeat was inserted.
In addition to the missense mutation described, we found an additional mutation associated with lin-36(n3765). This mutation, AG/gtaagaagaaaagc to AG/gtaagaagaaaagt, is present in the third intron of lin-36 and creates a possible splice-donor sequence. If this splice-donor were used, an in-frame ochre (TAA) stop codon would be encountered, truncating the LIN-36 protein after 261 amino acids.
These data are from Figure 3 of Thomas et al. (2003).
These data are from Figure 2 of Ceol and Horvitz (2004).
These data are from Figure 1 of Ceol and Horvitz (2004).
). The 41 include all of the dpl-1, efl-1, let-418, lin-9, lin-13, lin-36, lin-52, lin-53, lin-65, mep-1, mys-1, sli-1, and trr-1 alleles and one of two lin-35 alleles identified in our screen. Forty of 45 mutations are GC-to-AT transitions, which are characteristic of EMS mutagenesis (Anderson 1995). Many of these mutations are predicted to truncate the corresponding synMuv proteins. The truncations predicted by efl-1(n3639), let-418(n3719), lin-52(n3718), and trr-1(n3704) are particularly severe, and the synMuv and sterile abnormalities caused by these mutations likely represent the null phenotypes of these genes. In addition, we found missense mutations that disrupt predicted functional domains of synMuv proteins. For example, n3536, n3626, n3629, and one of the two mutations of n3636 affect the ATPase/helicase domain of LET-418. LET-418 is a member of the Mi-2 family of ATP-dependent chromatin remodeling enzymes (Solari and Ahringer 2000; von Zelewsky et al. 2000), and the LET-418 missense mutations suggest that LET-418 function is dependent on ATP hydrolysis. At least one mutation affecting the LIN-13 protein, n3642, is predicted to disrupt a canonical zinc-finger motif. This missense mutation, along with those isolated previously (Thomas et al. 2003), indicates that at least some of the 24 LIN-13 zinc fingers are important for LIN-13 synMuv activity. Missense mutations affecting other synMuv proteins are not as easily linked to the disruption of predicted functional domains. These mutations may provide useful starting points for identifying functional motifs within synMuv proteins that are not predicted by sequence comparisons.
DISCUSSION
Frequency of mutant isolation:
The rate at which we isolated synMuv mutations was much higher than that observed in previous screens. Considering screens that were conducted in class A synMuv mutant backgrounds, we recovered one synMuv mutation per 109 haploid genomes screened as compared with one per 750 (Ferguson and Horvitz 1989), one per 400 (Thomas et al. 2003), and one per 667 (Thomas et al. 2003) in previous screens. We believe the reasons for this difference are threefold. First, our screen design allowed the isolation of synMuv mutations that also caused sterility. Numerous sterile synMuv mutants had been observed in previous screens, but in general the mutations responsible were not recovered. Second, our parental strain carried a strong class A mutation, lin-15A(n767). The penetrance of the Muv phenotype of a synMuv strain is dependent on the combined strengths of the individual synMuv mutations (C. J. Ceol and H. R. Horvitz, unpublished observations). Therefore, even weak mutations could be identified in a strong synMuv background such as lin-15A(n767). Such weak mutations may not have been recovered in the three previous screens described above, all of which were performed in partial loss-of-function synMuv backgrounds. Third, by screening petri plates with many F2 progeny derived from a single F1 animal, we observed many genotypically identical animals for each haploid genome screened. Such screening can efficiently recover partially penetrant synMuv mutations.
Of the 62 mutations described in this study, 24 caused recessive sterility. The 38 mutations that did not cause sterility were recovered at 1 mutation per 178 haploid genomes screened, a frequency higher than that of previous screens. The difference in the rate of recovery of nonsterile mutants is likely a consequence of the second and third differences in screening described above.
Given that the average gene mutates to loss of function at a rate of ∼5 × 10−4 under the conditions of EMS mutagenesis we used (Brenner 1974; Meneely and Herman 1979; Greenwald and Horvitz 1980), our observed rate of 10−2 suggests that ∼20 genes can mutate by loss of function to cause a synMuv phenotype in combination with a class A synMuv mutation. Including the genes we identified in this study, a total of 25 such genes have been described to date. Three or fewer alleles of 15 of these genes have been recovered in synMuv screens, indicating that screens for such genes are not saturated.
Different synMuv gene classes likely act in parallel to antagonize let-60 Ras pathway activity:
Class A synMuv mutations synergize with class B mutations but not with other class A mutations, whereas class B synMuv mutations synergize with class A synMuv mutations but not with other class B synMuv mutations. Such genetic behavior led to the hypothesis that the A and B classes of synMuv genes encode components of two functionally redundant pathways that negatively regulate vulval development (Ferguson and Horvitz 1989). Consistent with this hypothesis, a subset of class B synMuv gene products has been shown to physically interact and their homologs are known to function together in other organisms (Lu and Horvitz 1998; Ceol and Horvitz 2001; Unhavaithaya et al. 2002; Korenjak et al. 2004; Lewis et al. 2004).
Because we conducted our screen using a class A synMuv background, we anticipated recovering mutations that affected class B synMuv genes. Indeed, 47 of the 62 mutations we isolated affected previously known and newly described class B synMuv genes. However, we discovered that some new mutations define new classes of synMuv genes. synMuv mutations previously were categorized by testing for synergism with class A and class B mutations. From such tests we discovered that some of our new mutations synthetically interacted with both class A and class B mutations; such mutations defined the class C genes trr-1 and mys-1 (this study and Ceol and Horvitz 2004). Other new mutations interacted like class B mutations in these standard tests but were distinguished from class B mutations by additional tests. For example, like class B mutations sli-1(n3538) synthetically interacted with class A but not with class B mutations yet, unlike class B mutations synthetically interacted with ark-1 and gap-1 and suppressed the let-23 Vul phenotype. These results led us to adopt two criteria when classifying synMuv mutations: (1) If two mutations synthetically interact to cause a Muv phenotype, then they are in different classes, and (2) if two mutations do not synthetically interact but interact differently with other classes of synMuv mutations or with let-23, then they are in different classes. Since we have found that interaction tests with only class A and class B mutations are insufficient to classify some synMuv genes, we suggest that previously described synMuv genes should be tested more extensively to establish their classifications.
Using more extensive genetic interaction tests and additional criteria to interpret these interactions, we define six classes of genes, synMuv A, synMuv B, synMuv C, gap-1, sli-1, and ark-1, that seem to act in parallel to each other to negatively regulate Ras-mediated vulval development (Table 9
Summary of synMuv genetic interactions
. | Phenotype of double mutant with mutation of specified class . | Phenotype of double mutant with let-23 . | Inferred synMuv class of mutant gene . | |||||
|---|---|---|---|---|---|---|---|---|
| Mutation . | Class A . | Class B . | Class C . | gap-1 . | sli-1 . | ark-1 . | ||
| lin-15A(n767) lin-38(n751) | non-Muv | Muv | Muv | Muv | Muv | Muv | Vul | A |
| lin-15B(n744) lin-35(n745) lin-65(n3441) mep-1(n3703)a | Muv | Non-Muv | Muv | Non-Muv | Non-Muv | Non-Muv | Vul | B |
| lin(n3628) | Muv | Non-Muv | Non-Muv | Non-Muv | Non-Muv | Non-Muv | Vul | B or lin(n3628)b |
| trr-1(n3712) | Muv | Muv | Non-Muv | Muv | Muv | Muv | Vul | C |
| gap-1(ga133) | Muv | Non-Muv | Muv | NA | Muv | Muv | Non-Vul | gap-1 |
| sli-1(n3538) | Muv | Non-Muv | Muv | Muv | NA | Muv | Non-Vul | sli-1 |
| ark-1(n3701) | Muv | Non-Muv | Muv | Muv | Muv | NA | Vul | ark-1 |
. | Phenotype of double mutant with mutation of specified class . | Phenotype of double mutant with let-23 . | Inferred synMuv class of mutant gene . | |||||
|---|---|---|---|---|---|---|---|---|
| Mutation . | Class A . | Class B . | Class C . | gap-1 . | sli-1 . | ark-1 . | ||
| lin-15A(n767) lin-38(n751) | non-Muv | Muv | Muv | Muv | Muv | Muv | Vul | A |
| lin-15B(n744) lin-35(n745) lin-65(n3441) mep-1(n3703)a | Muv | Non-Muv | Muv | Non-Muv | Non-Muv | Non-Muv | Vul | B |
| lin(n3628) | Muv | Non-Muv | Non-Muv | Non-Muv | Non-Muv | Non-Muv | Vul | B or lin(n3628)b |
| trr-1(n3712) | Muv | Muv | Non-Muv | Muv | Muv | Muv | Vul | C |
| gap-1(ga133) | Muv | Non-Muv | Muv | NA | Muv | Muv | Non-Vul | gap-1 |
| sli-1(n3538) | Muv | Non-Muv | Muv | Muv | NA | Muv | Non-Vul | sli-1 |
| ark-1(n3701) | Muv | Non-Muv | Muv | Muv | Muv | NA | Vul | ark-1 |
We provisionally assign 29 genes to six synMuv classes. The assignments of 11 of these genes (shown above and underlined below) are based on extensive genetic interaction tests. The remaining 18 genes have not been tested as extensively. However, on the basis of known genetic interactions and molecular identities, we speculate that most of these 18 genes will remain in the classes to which they have previously been assigned.
Class A: lin-8, lin-15A, lin-38, lin-56.
Class B: lin-9, lin-13, lin-15B, lin-35, lin-36, lin-37, lin-52, lin-53, lin-54, lin-61, lin-65, dpl-1, efl-1, hda-1, hpl-2, let-418, lin(n3628), mep-1.
Class C: trr-1, mys-1, epc-1, ssl-1.
gap-1: gap-1.
sli-1: sli-1.
ark-1: ark-1.
NA, not applicable; since each of these classes contains only one gene, double mutants within the same class cannot be constructed.
mep-1(n3703) and class C synMuv mutations interact to cause larval lethality at a stage earlier than vulval abnormalities can be determined.
Like class B synMuv mutations, lin(n3628) interacts synthetically with class A mutations; does not interact synthetically with class B, ark-1, gap-1, or sli-1 mutations; and does not suppress the Vul phenotype of let-23(sy97). However, lin(n3628) does not interact synthetically with class C mutations. lin(n3628) may define yet another class of synMuv genes. Alternatively, the mutation n3628 may be a partial loss-of-function mutation too weak to reveal redundancy with class C genes, in which case lin(n3628) may be a class B gene. Determination of the lin(n3628) null phenotype and genetic interaction tests with a null mutation of this gene should distinguish between these possibilities.
Summary of synMuv genetic interactions
. | Phenotype of double mutant with mutation of specified class . | Phenotype of double mutant with let-23 . | Inferred synMuv class of mutant gene . | |||||
|---|---|---|---|---|---|---|---|---|
| Mutation . | Class A . | Class B . | Class C . | gap-1 . | sli-1 . | ark-1 . | ||
| lin-15A(n767) lin-38(n751) | non-Muv | Muv | Muv | Muv | Muv | Muv | Vul | A |
| lin-15B(n744) lin-35(n745) lin-65(n3441) mep-1(n3703)a | Muv | Non-Muv | Muv | Non-Muv | Non-Muv | Non-Muv | Vul | B |
| lin(n3628) | Muv | Non-Muv | Non-Muv | Non-Muv | Non-Muv | Non-Muv | Vul | B or lin(n3628)b |
| trr-1(n3712) | Muv | Muv | Non-Muv | Muv | Muv | Muv | Vul | C |
| gap-1(ga133) | Muv | Non-Muv | Muv | NA | Muv | Muv | Non-Vul | gap-1 |
| sli-1(n3538) | Muv | Non-Muv | Muv | Muv | NA | Muv | Non-Vul | sli-1 |
| ark-1(n3701) | Muv | Non-Muv | Muv | Muv | Muv | NA | Vul | ark-1 |
. | Phenotype of double mutant with mutation of specified class . | Phenotype of double mutant with let-23 . | Inferred synMuv class of mutant gene . | |||||
|---|---|---|---|---|---|---|---|---|
| Mutation . | Class A . | Class B . | Class C . | gap-1 . | sli-1 . | ark-1 . | ||
| lin-15A(n767) lin-38(n751) | non-Muv | Muv | Muv | Muv | Muv | Muv | Vul | A |
| lin-15B(n744) lin-35(n745) lin-65(n3441) mep-1(n3703)a | Muv | Non-Muv | Muv | Non-Muv | Non-Muv | Non-Muv | Vul | B |
| lin(n3628) | Muv | Non-Muv | Non-Muv | Non-Muv | Non-Muv | Non-Muv | Vul | B or lin(n3628)b |
| trr-1(n3712) | Muv | Muv | Non-Muv | Muv | Muv | Muv | Vul | C |
| gap-1(ga133) | Muv | Non-Muv | Muv | NA | Muv | Muv | Non-Vul | gap-1 |
| sli-1(n3538) | Muv | Non-Muv | Muv | Muv | NA | Muv | Non-Vul | sli-1 |
| ark-1(n3701) | Muv | Non-Muv | Muv | Muv | Muv | NA | Vul | ark-1 |
We provisionally assign 29 genes to six synMuv classes. The assignments of 11 of these genes (shown above and underlined below) are based on extensive genetic interaction tests. The remaining 18 genes have not been tested as extensively. However, on the basis of known genetic interactions and molecular identities, we speculate that most of these 18 genes will remain in the classes to which they have previously been assigned.
Class A: lin-8, lin-15A, lin-38, lin-56.
Class B: lin-9, lin-13, lin-15B, lin-35, lin-36, lin-37, lin-52, lin-53, lin-54, lin-61, lin-65, dpl-1, efl-1, hda-1, hpl-2, let-418, lin(n3628), mep-1.
Class C: trr-1, mys-1, epc-1, ssl-1.
gap-1: gap-1.
sli-1: sli-1.
ark-1: ark-1.
NA, not applicable; since each of these classes contains only one gene, double mutants within the same class cannot be constructed.
mep-1(n3703) and class C synMuv mutations interact to cause larval lethality at a stage earlier than vulval abnormalities can be determined.
Like class B synMuv mutations, lin(n3628) interacts synthetically with class A mutations; does not interact synthetically with class B, ark-1, gap-1, or sli-1 mutations; and does not suppress the Vul phenotype of let-23(sy97). However, lin(n3628) does not interact synthetically with class C mutations. lin(n3628) may define yet another class of synMuv genes. Alternatively, the mutation n3628 may be a partial loss-of-function mutation too weak to reveal redundancy with class C genes, in which case lin(n3628) may be a class B gene. Determination of the lin(n3628) null phenotype and genetic interaction tests with a null mutation of this gene should distinguish between these possibilities.
). Some of these classes, such as gap-1, sli-1, and ark-1, likely interface directly with Ras pathway components (see below). The point at which the synMuv A, synMuv B, and synMuv C classes interface with Ras signaling is unknown, although recent studies suggest that the synMuv A and synMuv B classes may directly or indirectly repress inappropriate transcription of the Ras pathway activating ligand lin-3 in the syncytial hypodermis (Cui et al. 2006).
Different synMuv gene classes control distinct biochemical activities:
A synthetic genetic interaction implies functional redundancy between two sets of genes. There are many possible mechanisms by which two sets of genes might appear redundant. These possibilities include: (1) Two sets of genes encode similar sets of proteins with corresponding proteins of each set controlling the same biochemical activity, and hence each set controls the same biological process; (2) two sets of genes encode distinct sets of proteins with each set controlling distinct biochemical activities but the same biological process; and (3) two sets of genes encode distinct sets of proteins with each set regulating distinct but redundant biological processes.
The first of these mechanisms likely does not apply to the different classes of synMuv genes, as no cloned gene in one synMuv class is similar to any gene of another class. Furthermore, many of the cloned synMuv genes, including the class A gene lin-15A, the class B gene lin-35 Rb, the class C genes trr-1 and epc-1, and ark-1 and sli-1, encode the sole C. elegans member of their respective gene families.
The redundancy exhibited among sli-1, gap-1, and ark-1 likely exemplifies the second mechanism. sli-1, ark-1, and gap-1 are thought to directly downregulate Ras pathway activity, and, as might be predicted on the basis of their synthetic interactions, each is proposed to act upon a different Ras pathway component. sli-1 encodes a homolog of the c-Cbl proto-oncoprotein, which is thought to downregulate receptor tyrosine kinase levels though ubiquitin-mediated degradation (Yoon et al. 1995; Levkowitz et al. 1999). ark-1 encodes a protein that interacts with the SEM-5 SH2/SH3 adaptor protein and is predicted to be a cytoplasmic tyrosine kinase (Hopper et al. 2000). Since sem-5 acts downstream of the let-23 receptor tyrosine kinase, ark-1 is proposed to inhibit let-60 Ras signaling downstream of let-23. gap-1 is a member of the GTPase-activating protein (GAP) family (Hajnal et al. 1997). GAPs enhance the catalytic function of Ras family GTPases such as let-60 Ras, thereby facilitating the switch from active GTP-bound to inactive GDP-bound Ras. The genetic suppression of let-23(sy97) by and the molecular identities of sli-1 and gap-1 support their action downstream of the let-23 receptor tyrosine kinase. Although ark-1 mutations do not suppress let-23(sy97), Hopper et al. (2000) found that an ark-1 mutation suppressed the Vul phenotypes caused by weaker let-23 mutations and by sem-5 mutations. On the basis of these suppression data and the molecular data described above, these authors argued that ark-1 acts downstream of let-23, although its negative regulation of the let-60 pathway may not be as great as that of sli-1 or gap-1. The redundancy displayed by sli-1, gap-1, and ark-1 suggests that a mutation affecting one of these genes only mildly affects Ras pathway activity whereas mutations affecting two genes elevate pathway activity to a level that inappropriately transforms vulval cell fates. That these genes converge on the same signaling pathway implies that they regulate the same biological process.
The class A, B, and C synMuv genes may or may not act similarly. It is possible that these classes act on components of the let-60 Ras pathway. Since at least some class A and B synMuv genes are thought to act in the hypodermis, an effect on let-60 Ras signaling is likely indirect and may involve transcriptional regulation of the lin-3 ligand (Herman and Hedgecock 1990; Hedgecock and Herman 1995; Myers and Greenwald 2005; Cui et al. 2006). Alternatively, as in the case of the third mechanism, these classes may regulate entirely distinct biological processes. For example, the class B genes, some of which encode components of a putative histone deacetylase complex, may repress transcription of genes that indirectly promote P(3–8).p cell division. By contrast, the class C genes, which encode components of a putative histone acetyltransferase complex, may activate the transcription of genes, different from those targeted by class B genes, that promote differentiation of P(3–8).p descendants into hypodermal and not vulval cells. A better understanding of synMuv target genes should help to resolve whether different synMuv classes regulate the same or distinct biological activities.
Footnotes
Present address: Howard Hughes Medical Institute, Department of Hematology/Oncology, Children's Hospital, Boston, Massachusetts 02115.
Present address: Howard Hughes Medical Institute, Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115.
Footnotes
Communicating editor: B. J. Meyer
Acknowledgement
We thank Beth Castor, Na An, and Andrew Hellman for expert technical assistance and Erik Andersen and Adam Saffer for critical reading of this manuscript. We thank Yuji Kohara for providing cDNA clones, Mike Boxem and Sander van den Heuvel for providing pMB1 and pMB7, Min Han for providing pTG96, and Neil Hopper and Paul Sternberg for providing the ark-1(sy247) strain. Some of the strains used in this work were provided by Theresa Stiernagle of the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health (NIH) National Center for Research Resources. This work was supported by NIH grant GM24663 to H.R.H., a Koch graduate fellowship to C.J.C., and a Howard Hughes Medical Institute predoctoral fellowship to M.M.H. H.R.H. is the David H. Koch Professor of Biology at Massachusetts Institute of Technology and an Investigator of the Howard Hughes Medical Institute.
References
Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman,
Anderson, P.,
Aroian, R. V., and P. W. Sternberg,
Austin, J., and J. Kimble,
Beitel, G. J., S. G. Clark and H. R. Horvitz,
Beitel, G. J., E. J. Lambie and H. R. Horvitz,
Belfiore, M., L. D. Mathies, P. Pugnale, G. Moulder, R. Barstead et al.,
C. elegans Sequencing Consortium,
Ceol, C. J., and H. R. Horvitz,
Ceol, C. J., and H. R. Horvitz,
Chen, Z., and M. Han,
Clark, D. V., and D. L. Baillie,
Clark, S. G., A. D. Chisholm and H. R. Horvitz,
Clark, S. G., X. Lu and H. R. Horvitz,
Cui, M., J. Chen, T. R. Myers, B. J. Hwang, P. W. Sternberg et al.,
Davison, E. M., M. M. Harrison, A. J. Walhout, M. Vidal and H. R. Horvitz,
Dibb, N. J., D. M. Brown, J. Karn, D. G. Moerman, S. L. Bolten et al.,
Edgley, M. L., and D. L. Riddle,
Eisenmann, D. M., and S. K. Kim,
Ferguson, E. L., and H. R. Horvitz,
Ferguson, E. L., and H. R. Horvitz,
Ferguson, E. L., P. W. Sternberg and H. R. Horvitz,
Frankfort, B. J., and G. Mardon,
Freeman, M., and J. B. Gurdon,
Greenwald, I. S., and H. R. Horvitz,
Hajnal, A., C. W. Whitfield and S. K. Kim,
Han, M., and P. W. Sternberg,
Hedgecock, E. M., and R. K. Herman,
Hengartner, M. O., R. E. Ellis and H. R. Horvitz,
Herman, R. K.,
Herman, R. K., and E. M. Hedgecock,
Hill, R. J., and P. W. Sternberg,
Hodgkin, J.,
Hopper, N. A., J. Lee and P. W. Sternberg,
Hsieh, J., J. Liu, S. A. Kostas, C. Chang, P. W. Sternberg et al.,
Huang, L. S., P. Tzou and P. W. Sternberg,
Jongeward, G. D., T. R. Clandinin and P. W. Sternberg,
Kimble, J.,
Korenjak, M., B. Taylor-Harding, U. K. Binne, J. S. Saterlee, O. Stevaux et al.,
Levkowitz, G., H. Waterman, S. A. Ettenberg, M. Katz, A. Y. Tsygankov et al.,
Lewis, P. W., E. L. Beall, T. C. Fleischer, D. Georlette, A. J. Link et al.,
Lu, X.,
Lu, X., and H. R. Horvitz,
Maloof, J. N., and C. Kenyon,
Melendez, A., and I. Greenwald,
Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros,
Meneely, P. M., and R. K. Herman,
Moghal, N., and P. W. Sternberg,
Myers, T. R., and I. Greenwald,
Page, B. D., S. Guedes, D. Waring and J. R. Priess,
Poulin, G., Y. Dong, A. G. Fraser, N. A. Hopper and J. Ahringer,
Reddy, K. C., and A. M. Villeneuve,
Robinett, C. C., A. Straight, G. Li, C. Wilhelm, G. Sudlow et al.,
Rogalski, T. M., D. G. Moerman and D. L. Baillie,
Rosenbluth, R. E., and D. L. Baillie,
Sigurdson, D. C., G. J. Spanier and R. K. Herman,
Solari, F., and J. Ahringer,
Sternberg, P. W., and H. R. Horvitz,
Sulston, J. E., and H. R. Horvitz,
Thomas, J. H., and H. R. Horvitz,
Thomas, J. H., C. J. Ceol, H. T. Schwartz and H. R. Horvitz,
Unhavaithaya, Y., T. H. Shin, N. Miliaras, J. Lee, T. Oyama et al.,
von Zelewsky, T., F. Palladino, K. Brunschwig, H. Tobler, A. Hajnal et al.,
Wang, B. B., M. M. Muller-Immergluck, J. Austin, N. T. Robinson, A. Chisholm et al.,
Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk,
Williams, B. D., B. Schrank, C. Huynh, R. Shownkeen and R. H. Waterston,
Yoon, C. H., J. Lee, G. D. Jongeward and P. W. Sternberg,
Zorio, D. A., N. N. Cheng, T. Blumenthal and J. Spieth,