-
PDF
- Split View
-
Views
-
Cite
Cite
Ambro van Hoof, Conserved Functions of Yeast Genes Support the Duplication, Degeneration and Complementation Model for Gene Duplication, Genetics, Volume 171, Issue 4, 1 December 2005, Pages 1455–1461, https://doi.org/10.1534/genetics.105.044057
Close - Share Icon Share
Abstract
Gene duplication is often cited as a potential mechanism for the evolution of new traits, but this hypothesis has not been thoroughly tested experimentally. A classical model of gene duplication states that after gene duplication one copy of the gene preserves the ancestral function, while the other copy is free to evolve a new function. In an alternative duplication, divergence, and complementation model, duplicated genes are preserved because each copy of the gene loses some, but not all, of its functions through degenerating mutations. This results in the degenerating mutations in one gene being complemented by the other and vice versa. These two models make very different predictions about the function of the preduplication orthologs in closely related species. These predictions have been tested here for several duplicated yeast genes that appeared to be the leading candidates to fit the classical model. Surprisingly, the results show that duplicated genes are maintained because each copy carries out a subset of the conserved functions that were already present in the preduplication gene. Therefore, the results are not consistent with the classical model, but instead fit the duplication, divergence, and complementation model.
THE mechanisms by which an organism acquires new traits have been discussed since before Darwin's On the Origin of Species, and gene duplication is often cited as one potential mechanism for the evolution of new traits. The “classical model” for the role of gene duplication in the evolution of new functions states that after gene duplication, two genes that carry out the same functions are redundant. This could result in one gene maintaining that function, leaving the other gene free to mutate and occasionally evolve a new function (Ohno 1970). Ohno further predicted that duplicated pairs of genes would evolve at different rates, with a slowly evolving copy maintaining the original function and a fast-evolving copy gaining a new function. This model has recently been widely cited because these predictions appear to fit some, but not all, cases of the maintenance of duplicated genes after whole genome duplication (e.g., Andalis et al. 2004; Goffeau 2004; Jaillon et al. 2004; Kellis et al. 2004; Piskur and Langkjaer 2004; Wolfe 2004). However, there is limited direct experimental evidence that gene duplication allows evolution of new functions.
Alternative models for the fate of duplicated genes are referred to as duplication, degeneration, and complementation (DDC) models (e.g., Hughes 1994; Force et al. 1999). They state that many genes have two (or more) distinct functions. After duplication, one copy is free to mutate so that it no longer carries out one function, and the other copy is free to mutate so that it no longer carries out the other function. Thus, the two degenerated genes complement each other and together carry out the same two functions as the preduplication gene. The key difference between the classical model and DDC models is that in the classical model gene duplication precedes (and allows) the evolution of a new function, while in DDC models the ancestral gene evolved a second function before duplication, and duplicated genes are maintained because they each lost one or more functions.
One variation of DDC models states that a preduplication gene that is expressed under two conditions can evolve into two genes that are each expressed under one of those conditions. Some experimental evidence supports this differential expression variant of DDC models. For example, the mouse and chicken have an en1 gene that is expressed in the pectoral appendage bud and in some neurons, while zebrafish have one ortholog that is expressed in the pectoral appendage bud and a second ortholog that is expressed in neurons (Force et al. 1999). Similarly, Arabidopsis has an AGAMOUS gene that is expressed in developing carpels and stamens, while maize has two genes with one of them expressed in developing carpels and the other in developing stamens (Force et al. 1999).
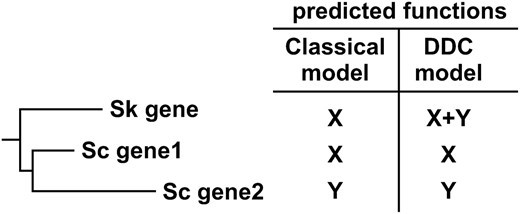
Approximately 100–300 MYA the whole genome of an ancestor of the yeast Saccharomyces cerevisiae was duplicated (Wolfe and Shields 1997; Friedman and Hughes 2001; Kellis et al. 2004). After genome duplication, one copy of most pairs of genes was lost, but some pairs of duplicated genes have been preserved. Such genome duplication and subsequent loss of one member of each gene pair is not unique to yeast, but also occurred in fish and in rice (Jaillon et al. 2004; Yu et al. 2005). However, yeast offers many experimental tools that can be used to study the function of duplicated genes. S. kluyveri diverged from S. cerevisiae before whole-genome duplication, and thus functions common between S. kluyveri and S. cerevisiae orthologs are most likely functions that were already present in the preduplication ancestral yeast gene. Therefore, the classical model specifically predicts that for any duplicated pair of S. cerevisiae genes, the orthologous S. kluyveri gene may complement a mutation in the slower-evolving S. cerevisiae ortholog, but not in the faster-evolving S. cerevisiae ortholog (Figure 1). In contrast, DDC models predict that S. kluyveri genes may complement mutations in both S. cerevisiae orthologs, since they state that the preduplication gene carried out both functions.
The classical model of gene duplication and duplication, degeneration, and complementation (DDC) models make distinct predictions about what functions are conserved in the S. kluyveri (Sk) ortholog of duplicated S. cerevisiae (Sc) genes.
The experiments presented here test the above prediction for a set of candidates most likely to fit the classical model. The results indicate that these candidates do not fit the classical model, but instead fulfill the predictions of the DDC model. Further analysis of one of the duplicated gene pairs, and previously published functional analysis of other gene pairs, suggests that these gene pairs do not fit the differential expression variant of the DDC model, but instead fit the DDC model with the loss-of-function mutations having occurred within the coding region.
MATERIALS AND METHODS
S. kluyveri was used as a source of preduplication orthologs, because among the sequenced yeast genomes it and Kluyveromyces waltii are the closest preduplication relatives of S. cerevisiae. S. kluyveri appears to have evolved more slowly than K. waltii (Kurtzman and Robnett 2003), which may increase the chance that its genes would complement S. cerevisiae mutants. S. kluyveri orthologs of duplicated S. cerevisiae genes were identified by conserved synteny and by sequence similarity (using BLAST).
The S. kluyveri strain FM479 was obtained from Mark Johnston (Washington University, St. Louis). PCR primers were designed to amplify S. kluyveri genes, including ∼500 bp of their native promoters and ∼500 bp of their native 3′-UTRs (Table 1). The PCR product was cloned into the low-copy centromeric plasmid pRS416 (Sikorski and Hieter 1989) using restriction enzyme sites in the PCR primers. The 5′- and 3′-end of the S. kluyveri SIR3/ORC1 ortholog are present in different contigs in GenBank, but the whole gene was successfully amplified from genomic DNA and the gap was sequenced, thus confirming that these two contigs are adjacent to each other in the S. kluyveri genome. The HBS1/SKI7 gene of S. kluyveri contains a putative intron (data not shown), which is included in the complementing plasmid.
Oligonucleotides used
Name . | Description . | Sequence . |
|---|---|---|
| oAV144 | 5′ Sk HBS1/SKI7 | cgagaattcggtggtagaacgttgtacaagg |
| oAV145 | 3′ Sk HBS1/SKI7 | cgatctagacaatagaaaccacacaagagg |
| oAV201 | 5′ Sk ORC1/SIR3 | cgacgactagtggaagtcctgtacatctttgagtacc |
| oAV202 | 3′ Sk ORC1/SIR3 | cgacggcggccgcccctgctcatttataagtcccttg |
| oAV207 | 5′ Sk RNR2/RNR4 | cgatctagatctcaaacccagccaagcc |
| oAV208 | 3′ Sk RNR2/RNR4 | cgactcgagatgtccatgccacttgcacg |
| oAV209 | 5′ Sk SNF12/RSC6 | cgatctagaggtacgggatcaaacagctg |
| oAV210 | 3′ Sk SNF12/RSC6 | cgactcgagacgtatgtagtgatgcgagg |
| oAV177 | 5′ TDH-SKI7 | cgaactagtaccacctgacatgtcgttattag |
| oAV178 | 3′ TDH-SKI7 | cgactcgagggattactggcatgcaattctg |
| oAV179 | 5′ TDH-HBS1 | cgatctagactatcgagatggcttacagtg |
| oAV180 | 3′ TDH-HBS1 | cgactcgagcccttctactgagttatttcgg |
Name . | Description . | Sequence . |
|---|---|---|
| oAV144 | 5′ Sk HBS1/SKI7 | cgagaattcggtggtagaacgttgtacaagg |
| oAV145 | 3′ Sk HBS1/SKI7 | cgatctagacaatagaaaccacacaagagg |
| oAV201 | 5′ Sk ORC1/SIR3 | cgacgactagtggaagtcctgtacatctttgagtacc |
| oAV202 | 3′ Sk ORC1/SIR3 | cgacggcggccgcccctgctcatttataagtcccttg |
| oAV207 | 5′ Sk RNR2/RNR4 | cgatctagatctcaaacccagccaagcc |
| oAV208 | 3′ Sk RNR2/RNR4 | cgactcgagatgtccatgccacttgcacg |
| oAV209 | 5′ Sk SNF12/RSC6 | cgatctagaggtacgggatcaaacagctg |
| oAV210 | 3′ Sk SNF12/RSC6 | cgactcgagacgtatgtagtgatgcgagg |
| oAV177 | 5′ TDH-SKI7 | cgaactagtaccacctgacatgtcgttattag |
| oAV178 | 3′ TDH-SKI7 | cgactcgagggattactggcatgcaattctg |
| oAV179 | 5′ TDH-HBS1 | cgatctagactatcgagatggcttacagtg |
| oAV180 | 3′ TDH-HBS1 | cgactcgagcccttctactgagttatttcgg |
Oligonucleotides used
Name . | Description . | Sequence . |
|---|---|---|
| oAV144 | 5′ Sk HBS1/SKI7 | cgagaattcggtggtagaacgttgtacaagg |
| oAV145 | 3′ Sk HBS1/SKI7 | cgatctagacaatagaaaccacacaagagg |
| oAV201 | 5′ Sk ORC1/SIR3 | cgacgactagtggaagtcctgtacatctttgagtacc |
| oAV202 | 3′ Sk ORC1/SIR3 | cgacggcggccgcccctgctcatttataagtcccttg |
| oAV207 | 5′ Sk RNR2/RNR4 | cgatctagatctcaaacccagccaagcc |
| oAV208 | 3′ Sk RNR2/RNR4 | cgactcgagatgtccatgccacttgcacg |
| oAV209 | 5′ Sk SNF12/RSC6 | cgatctagaggtacgggatcaaacagctg |
| oAV210 | 3′ Sk SNF12/RSC6 | cgactcgagacgtatgtagtgatgcgagg |
| oAV177 | 5′ TDH-SKI7 | cgaactagtaccacctgacatgtcgttattag |
| oAV178 | 3′ TDH-SKI7 | cgactcgagggattactggcatgcaattctg |
| oAV179 | 5′ TDH-HBS1 | cgatctagactatcgagatggcttacagtg |
| oAV180 | 3′ TDH-HBS1 | cgactcgagcccttctactgagttatttcgg |
Name . | Description . | Sequence . |
|---|---|---|
| oAV144 | 5′ Sk HBS1/SKI7 | cgagaattcggtggtagaacgttgtacaagg |
| oAV145 | 3′ Sk HBS1/SKI7 | cgatctagacaatagaaaccacacaagagg |
| oAV201 | 5′ Sk ORC1/SIR3 | cgacgactagtggaagtcctgtacatctttgagtacc |
| oAV202 | 3′ Sk ORC1/SIR3 | cgacggcggccgcccctgctcatttataagtcccttg |
| oAV207 | 5′ Sk RNR2/RNR4 | cgatctagatctcaaacccagccaagcc |
| oAV208 | 3′ Sk RNR2/RNR4 | cgactcgagatgtccatgccacttgcacg |
| oAV209 | 5′ Sk SNF12/RSC6 | cgatctagaggtacgggatcaaacagctg |
| oAV210 | 3′ Sk SNF12/RSC6 | cgactcgagacgtatgtagtgatgcgagg |
| oAV177 | 5′ TDH-SKI7 | cgaactagtaccacctgacatgtcgttattag |
| oAV178 | 3′ TDH-SKI7 | cgactcgagggattactggcatgcaattctg |
| oAV179 | 5′ TDH-HBS1 | cgatctagactatcgagatggcttacagtg |
| oAV180 | 3′ TDH-HBS1 | cgactcgagcccttctactgagttatttcgg |
To express S. cerevisiae Ski7p and Hbs1p under control of the TDH3 promoter, the coding regions were PCR amplified and cloned into p426GPD, using restriction enzyme sites in the PCR primers (Table 1; Mumberg et al. 1995).
Strain yRP1536 (dcp1-2 ski7Δ) (van Hoof et al. 2000) and strain TKY609 (rps30aΔ hbs1Δ) have been described (Carr-Schmid et al. 2002). All other strains were obtained from Open Biosystems (Huntsville, AL). Each strain was confirmed to carry the correct deletion by PCR. For ski7Δ, hbs1Δ, and sir3Δ strains the plasmids described above were introduced into haploid strains using standard yeast transformation protocols. For orc1Δ, rnr2Δ, rnr4Δ, snf12Δ, and rsc6Δ, the plasmids described above were introduced into heterozygous diploid strains. Haploid progeny spores were obtained by the hydrophobic spore isolation method essentially as described by Rockmill et al. and plated on YPD (Rockmill et al. 1991). The diploid strains are heterozygous for the knock out, lys2Δ, and met15Δ. At least 50 individual colonies were picked and scored for these segregating markers by replica plating onto YPD+geneticin, SC-MET-CYS, and SC-LYS. Haploid progeny carrying the deletion mutation were identified by their growth on YPD+geneticin and a failure to grow on SC-LYS and/or SC-MET-CYS media. These same 50 progeny strains were also replica plated to SC-URA to identify progeny that had inherited the plasmid and to 5-FOA media to identify progeny that were unable to lose the plasmid. All haploid deletion mutants grew on SC-URA, indicating that they had inherited the plasmid, and were unable to grow on 5-FOA plates, indicating that they were unable to lose this plasmid. In each case results from one such progeny are shown, but the other progeny showed the same results.
To assay mating capability, the indicated sir3Δ or wild-type control strains that all are MATa, HIS1, and his3 were grown with a MATα, his1, HIS3 strain (DTY8) on SC-URA plates. After overnight growth and mating, the mixture was serially diluted in fivefold increments and spotted onto the minimal medium plate shown in Figure 2 using an 8 × 6 array replica plater (Sigma, St. Louis). Only resulting diploids can grow on this minimal medium. For all other assays, the indicated strains were grown, serially diluted in fivefold increments, and plated on the indicated media. Plates were incubated at either 25° or 30° (unless otherwise indicated) and growth was monitored for several days.
RESULTS
Specific duplicated genes to be tested were identified on the basis of five criteria. First, to ensure that the genes studied resulted from duplication of the whole gene, a list of paralogs that arose by genome duplication was used as a starting point (Kellis et al. 2004). These ∼450 gene pairs were identified on the basis of a pattern of “double conserved synteny” that is characteristic of whole-genome duplication. Second, gene pairs with one or two genes of unknown function were excluded. For example, both the paralogs yCL069W and yKR105C have unknown functions, making it impossible to determine which functions are conserved or newly evolved. Similarly, gene pairs with largely overlapping function were excluded. For example, TEF1 and TEF2 encode proteins with the exact same amino acid sequence that appears functionally interchangeable (Carr-Schmid et al. 1999). Third, one specific prediction of the classical model is that gene duplication results in one more rapidly evolving paralog and one more slowly evolving paralog. Gene pairs that fit this prediction were chosen for characterization of conserved vs. newly evolved functions. Fourth, gene pairs in which overexpression of one paralog can complement deletion of the other paralog were excluded. This was done because while S. kluyveri orthologs were introduced with their native promoter on a low-copy plasmid, it is impossible to ensure that the introduced S. kluyveri gene is not overexpressed. Fifth, for practical reasons gene pairs that are well characterized and have easily assayable mutant phenotypes as described in the Saccharomyces genome database (http://www.yeastgenome.org) were chosen for characterization.
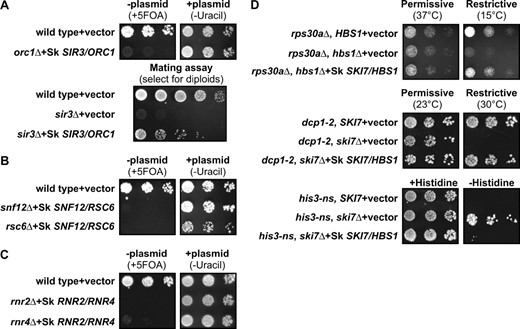
One pair of duplicated S. cerevisiae genes that fits the criteria specified above consists of ORC1 and SIR3. The Orc1p protein is essential for viability and is part of the origin recognition complex that functions during DNA replication (Bell et al. 1995). Sir3p is also part of a protein complex that includes some, but not all, of the same proteins as the Orc1p complex. The Sir3p complex functions in silencing genes near telomeres and at the silent mating-type loci. As a result, mutants lacking Sir3p are alive, but cannot mate (Rine and Herskowitz 1987). Additional copies of the SIR3 gene cannot complement an orc1 mutation and vice versa, further indicating that the proteins carry out different functions (Bell et al. 1995). The S. kluyveri genome encodes one gene that is 48% identical to Orc1p and 24% identical to Sir3p (see supplemental material at http://www.genetics.org.supplemental/). Thus, Orc1p evolved more slowly after gene duplication and the classical model predicts that Orc1p carries out a conserved function, while Sir3p carries out a newly evolved function (Kellis et al. 2004). To directly test which of these two functions is ancestral, the S. kluyveri ORC1/SIR3 gene was cloned onto a low-copy plasmid and introduced into S. cerevisiae deletion mutants lacking either ORC1 or SIR3. As expected by both models, the S. kluyveri gene restored viability to the orc1Δ mutant (Figure 2A). Importantly, the S. kluyveri gene also restored mating ability to the sir3Δ mutant. Thus, the S. kluyveri protein is capable of carrying out both Orc1p and Sir3p functions. The most parsimonious explanation of these data is that the two S. cerevisiae genes evolved from an ancestral gene that carried out two distinct and separable functions.
The single S. kluyveri ortholog of duplicated S. cerevisiae genes can complement complete deletions of both genes, consistent with duplication, degeneration, and complementation models. (A–C, top) Growth of the orc1Δ, snf12Δ, rsc6Δ, rnr2Δ, and rnr4Δ mutant strains that contain the indicated plasmid in the presence of 5-FOA or the absence of uracil is shown. 5-FOA selects against the plasmid (−plasmid), and failure to grow under these conditions shows the mutant phenotype (left). These mutant phenotypes are complemented by the respective S. kluyveri orthologs as indicated by the growth in the absence of uracil, which selects for the plasmid carrying the S. kluyveri gene (+plasmid). (A, bottom) Only diploids formed after successful mating can grow on the minimal medium plate shown. The sir3Δ mutant is unable to mate and form diploids (middle row), and this defect is complemented by the S. kluyveri gene (bottom row). (D, top) hbs1Δ rps30aΔ double mutants grow slowly at 15°, and this defect is complemented by the S. kluyveri ortholog. (Middle) ski7 dcp1-2 double mutants are inviable at 30°. dcp1-2 is a temperature-sensitive mutation that inactivates the mRNA decapping enzyme at 30°. This defect is complemented by the S. kluyveri SKI7/HBS1 ortholog. (Bottom) ski7Δ his3-nonstop double mutants grow in the absence of histidine, and this growth phenotype is complemented by the S. kluyveri ortholog.
A second pair of duplicated S. cerevisiae genes encodes Snf12p and Rsc6p, which are subunits of different chromatin remodeling complexes. Specifically, Snf12p is a subunit of the SWI/SNF complex that regulates transcription of the SUC2 and HO endonuclease genes as well as other genes. (Cairns et al. 1996a). Rsc6p is a subunit of the related Rsc complex that regulates transcription of ribosomal protein genes as well as other genes (Cairns et al. 1996b; Angus-Hill et al. 2001). Overexpression of Snf12p or Rsc6p does not complement a mutation in the other gene, even when introduced on a high-copy plasmid, indicating that they carry out distinct functions (Cairns et al. 1996b). The S. kluyveri genome encodes only one protein that is 37% identical to Snf12p and 28% identical to Rsc6p (see supplemental material at http://www.genetics.org/supplemental/). Thus, the classical model predicts that Snf12p carries out an ancestral function and Rsc6p carries out a newly evolved function. In the strain used here, SNF12 is an essential gene, and expression of the single S. kluyveri gene restores growth to the snf12Δ strain (Figure 2B). The faster-evolving paralog RSC6 is also an essential gene, and the S. kluyveri SNF12/RSC6 gene also restored viability to the rsc6Δ strain. The most parsimonious explanation of these data is that the preduplication ancestral gene carried out both Snf12p and Rsc6p functions.
A third example of duplicated S. cerevisiae genes are the two R2 subunits of ribonucleotide reductase (RNR) that are encoded by the RNR2 and RNR4 genes (Elledge and Davis 1987; Huang and Elledge 1997; Wang et al. 1997; Kellis et al. 2004). S. kluyveri contains only one RNR R2 subunit gene that is orthologous to both RNR2 and RNR4. Rnr2p is essential for viability and 82% identical to the S. kluyveri protein (Elledge and Davis 1987). Rnr4p is the more rapidly evolving paralog (48% identity) and deletion of RNR4 results in slow growth under a variety of conditions in some strains and inviability in other strains (see supplemental material at http://www.genetics.org/supplemental/; Huang and Elledge 1997; Wang et al. 1997). These phenotypes cannot be complemented by multiple copies of the other gene, even when present on a high-copy plasmid, suggesting that both have distinct functions within the RNR complex (Huang and Elledge 1997; Wang et al. 1997). Figure 2C shows that the single S. kluyveri RNR gene complements both the rnr2Δ and the rnr4Δ mutation. Thus, the functions of Rnr2p and Rnr4p were most likely encoded in one preduplication gene.
A fourth example of a pair of duplicated S. cerevisiae genes is HBS1 and SKI7. The genome of S. kluyveri encodes one protein that is 47% identical to Hbs1p and 16% identical to Ski7p (see supplemental material at http://www.genetics.org/supplemental/). Thus the classical model predicts that Hbs1p carries out an ancestral function, while Ski7p has a newly evolved function. The function of Hbs1p is not well defined, but lack of Hbs1p causes a slow growth phenotype at low temperature in strains that also lack the RPS30A gene (Carr-Schmid et al. 2002). As shown in Figure 2D, the HBS1/SKI7 gene from S. kluyveri is capable of complementing this phenotype of a hbs1Δ rps30aΔ double mutant. The paralog Ski7p functions in mRNA degradation, and mutants lacking SKI7 have three known phenotypes. First, yeast has two general pathways to degrade mRNA. As a result, mutants lacking Ski7p are viable in an otherwise wild-type strain, but inviable if the second mRNA decay pathway is also disabled (van Hoof et al. 2000). This inviability can be made conditional by using a temperature-sensitive mutation (such as dcp1-2) to inactivate the second pathway (van Hoof et al. 2000). Figure 2D shows that the S. kluyveri HBS1/SKI7 gene can complement this conditional inviability. Second, mRNAs that lack a stop codon are preferentially degraded by the Ski7p-dependent pathway. Therefore, a his3-nonstop ski7Δ double mutant grows better in the absence of histidine than a his3-nonstop SKI7 strain (van Hoof et al. 2002). Figure 2D shows that the S. kluyveri HBS1/SKI7 gene can also complement this phenotype. Third, ski7 mutations were initially identified because they have a super killer phenotype. That is, ski7 mutants that carry the killer virus secrete more killer toxin, and are more effective at killing uninfected yeast, than the wild-type strain (Ridley et al. 1984; Benard et al. 1999). This phenotype is also complemented by the S. kluyveri HBS1/SKI7 gene (data not shown). Thus, the S. kluyveri protein is capable of carrying out both Hbs1p and Ski7p functions. These data indicate the preduplication ancestral gene carried out both Ski7p and Hbs1p functions.
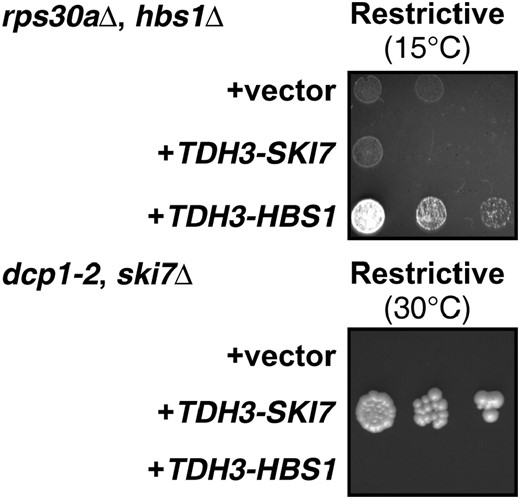
For the SKI7/HBS1 gene pair it is not known whether overexpression of one paralog can suppress deletion of the other. To directly test this, the coding region of either gene was put under control of the strong TDH3 promoter and CYC1 3′-UTR on a high-copy plasmid. As shown in Figure 3, the TDH3-SKI7 construct can complement ski7Δ but not hbs1Δ, and TDH3-HBS1 can complement hbs1Δ but not ski7Δ. These data confirm that Hbs1p and Ski7p have distinct functions.
Mutations in the coding regions are responsible for the subfunctionalization of SKI7 and HBS1. Experimental details are as in Figure 2.
DISCUSSION
The leading candidates for the classical model support the alternative DDC model:
The results presented here are inconsistent with the classical model for preservation of duplicated genes, even though one of the selection criteria for what genes to study was that they were among the best candidates to fit that model. Specifically, the classical model predicts that duplicated gene pairs consist of a slower-evolving gene with a conserved function and a faster-evolving gene with a new function. The genes studied here have widely varying levels of sequence identity with their S. kluyveri orthologs, but each duplicated gene pair studied included one more rapidly evolving paralog and one more slowly evolving paralog. In the case of Sir3p it was especially attractive to propose a new function in the silencing of mating-type cassettes, because these cassettes evolved at about the same time as the whole-genome duplication (Langkjaer et al. 2003; Butler et al. 2004). Surprisingly, the data presented here show that none of these candidates actually fit the classical model of gene duplication.
Although the results presented here do not provide any evidence for newly evolved functions, many proteins may have multiple functions, and any of the duplicated genes analyzed here may have acquired a new function, just like any of the nonduplicated yeast genes may also have acquired a new function. Importantly, the above results show that such hypothetical newly acquired functions are not required to explain why these duplicated genes were maintained. Most importantly, these results demonstrate that maintaining distinct ancestral functions provides sufficient evolutionary advantage to fully explain the maintenance of these duplicated genes. For example, the ORC1 gene is likely maintained because without its ancestral function yeast is inviable, and the SIR3 gene is likely maintained because without its ancestral function yeast is incapable of mating. Therefore, these results provide strong experimental support for the duplication, divergence, and complementation models.
Some support for the classical model for gene duplications has been found in two-hybrid analysis of protein-protein interaction in S. cerevisiae. Most recently, it was reported that the total number of two-hybrid interactions for a duplicated gene pair is larger than the number for a nonduplicated gene (He and Zhang 2005), suggesting that many protein interactions have been added after gene duplication. There are several possible reasons for the apparent discrepancy between those results and the results reported here. First, the experiments described here looked at a subset of gene duplications that arose by genome duplications as identified by a pattern of double-conserved synteny. The two-hybrid study identified paralogs by sequence similarity (BLAST). Paralogs identified by a high BLAST score may include proteins that arose by domain swapping. That is, part of a protein may have been duplicated and fused to another protein. The proteins studied here arose by whole-genome duplication and align along their entire length (see supplemental material at http://www.genetics.org/supplemental/; Kellis et al. 2004). While both complete and partial gene duplications should be considered, they probably should be considered separately. Second, analysis of protein interactions compared duplicated genes to nonduplicated genes within S. cerevisiae, whereas in the current analysis the duplicated genes were compared to their nonduplicated S. kluyveri ortholog. Comparing duplicated S. cerevisiae genes to nonduplicated S. cerevisiae genes would be invalid if there is an underlying bias as to what genes survived as duplicates. Bias toward maintaining certain genes in duplicate is evident from the fact that ∼75% of ribosomal protein genes are duplicated as compared to 12% of all genes (Warner et al. 2001; Kellis et al. 2004; and data not shown). Genes that have more separable functions also may be more likely to be maintained after gene duplication (Lynch and Force 2000; Walsh 2003).
DDC can occur by mutations within the coding region:
In the cases of ORC1/SIR3, RNR2/RNR4, and SKI7/HBS1 divergence of function is likely to have occurred through amino acid sequence changes. This conclusion is based on different lines of evidence for the three gene pairs. In the case of SIR3/ORC1, this hypothesis is based on characterization of chimaeric proteins. Specifically, a fusion protein that contains the C-terminal region of Sir3p and the N-terminal region of Orc1p can complement a sir3 mutation, but not an orc1 mutation, while a fusion protein that contains the C-terminal region of Orc1p and the N-terminal region of Sir3p can complement an orc1 mutation, but not a sir3 mutation (Bell et al. 1995). In the case of SKI7/HBS1, a chimaeric gene containing either the SKI7 or HBS1 coding region with heterologous promoters and 3′-UTRs can complement a deletion of the corresponding gene, but not a deletion of the paralog (Figure 3). Thus, in both SKI7/HBS1 and ORC1/SIR3 diverging mutations occurred within the coding region. The most likely explanation is that these gene pairs diverged through amino acid changes, although the possibility that some element within the coding region controls expression cannot be completely excluded. In the case of RNR2/RNR4, this hypothesis is based on the conclusion that Rnr2p and Rnr4p function as a heterodimer and thus that amino acid differences (and not expression differences) between Rnr2p and Rnr4p probably explain their different functions (Sommerhalter et al. 2004).
While previously published examples provided evidence for regulatory divergence (Force et al. 1999), the HBS1/SKI7, ORC1/SIR3, and RNR2/RNR4 gene pairs likely functionally diverged through amino acid changes. There are at least two explanations for this difference. One reason that most of the gene pairs studied here functionally diverged by amino acid changes is that they were selected in part because they included a fast-evolving paralog. Other yeast gene pairs, such as those that encode very similar proteins, may have diverged through changes in expression pattern. Second, functional divergence of expression patterns may be more prevalent for genes involved in development of multicellular organisms (such as en1 and AGAMOUS; Force et al. 1999), but less important in a unicellular microbe such as yeast. In either case, the results presented here offer direct evidence that functional divergence within the coding region has occurred during evolution and is important for the maintenance of duplicated genes.
In conclusion, four sets of paralogs in S. cerevisiae were analyzed. These four sets were carefully chosen because they were among the best candidates for the evolution of new function after yeast genome duplication. Contrary to the predictions of the classical model, the different functions of the paralogs were clearly already present in the preduplication ancestor. All of the data presented here are consistent with the hypothesis that the preservation of duplicated yeast genes can be explained by duplication, degeneration, and complementation and that newly evolved functions have contributed little to the persistence of most duplicated genes in yeast. These data are fully consistent with theoretical and population genetic analyses of DDC models (e.g., Lynch and Force 2000; Walsh 2003) and offer strong experimental support for such models. One limitation of the population genetic analyses is that they require assumptions about the relative rate of different kinds of mutations. Specifically, the ratio between mutations that lead to the loss of one function and mutations that lead to loss of all functions is an important, but unknown, variable in population genetic calculations. On the basis of the current work it should be possible to experimentally determine this ratio with the use of S. cerevisiae strains complemented by bifunctional S. kluyveri genes.
Footnotes
Communicating editor: A. Mitchell
Acknowledgement
I thank Roy Parker, Kevin Morano, and Aaron Mitchell for providing thoughtful comments on this manuscript. Kevin Morano generously supplied plasmid p426GPD and yeast strain TDY8. Mark Johnston generously supplied the S. kluyveri strain used. This work was supported by the PEW Scholarship Program in the Biomedical Sciences.
References
Andalis, A. A., Z. Storchova, C. Styles, T. Galitski, D. Pellman et al.,
Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst et al.,
Bell, S. P., J. Mitchell, J. Leber, R. Kobayashi and B. Stillman,
Benard, L., K. Carroll, R. C. Valle, D. C. Masison and R. B. Wickner,
Butler, G., C. Kenny, A. Fagan, C. Kurischko, C. Gaillardin et al.,
Cairns, B. R., R. S. Levinson, K. R. Yamamoto and R. D. Kornberg,
Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis et al.,
Carr-Schmid, A., N. Durko, J. Cavallius, W. C. Merrick and T. G. Kinzy,
Carr-Schmid, A., C. Pfund, E. A. Craig and T. G. Kinzy,
Elledge, S. J., and R. W. Davis,
Force, A., M. Lynch, F. B. Pickett, A. Amores, Y. L. Yan et al.,
Friedman, R., and A. L. Hughes,
He, X., and J. Zhang,
Huang, M., and S. J. Elledge,
Hughes, A. L.,
Jaillon, O., J. M. Aury, F. Brunet, J. L. Petit, N. Stange-Thomann et al.,
Kellis, M., B. W. Birren and E. S. Lander,
Kurtzman, C. P., and C. J. Robnett,
Langkjaer, R. B., P. F. Cliften, M. Johnston and J. Piskur,
Lynch, M., and A. Force,
Mumberg, D., R. Muller and M. Funk,
Piskur, J., and R. B. Langkjaer,
Ridley, S. P., S. S. Sommer and R. B. Wickner,
Rine, J., and I. Herskowitz,
Rockmill, B., E. J. Lambie and G. S. Roeder,
Sikorski, R. S., and P. Hieter,
Sommerhalter, M., W. C. Voegtli, D. L. Perlstein, J. Ge, J. Stubbe et al.,
van Hoof, A., R. R. Staples, R. E. Baker and R. Parker,
van Hoof, A., P. A. Frischmeyer, H. C. Dietz and R. Parker,
Walsh, B.,
Wang, P. J., A. Chabes, R. Casagrande, X. C. Tian, L. Thelander et al.,
Warner, J. R., J. Vilardell and J. H. Sohn,
Wolfe, K.,
Wolfe, K. H., and D. C. Shields,
Yu, J., J. Wang, W. Lin, S. Li, H. Li et al.,