-
PDF
- Split View
-
Views
-
Cite
Cite
Yuko Todaka, Yonggang Wang, Kosuke Tashiro, Nobutaka Nakashima, Takeharu Nishimoto, Takeshi Sekiguchi, Association of the GTP-Binding Protein Gtr1p With Rpc19p, a Shared Subunit of RNA Polymerase I and III in Yeast Saccharomyces cerevisiae, Genetics, Volume 170, Issue 4, 1 August 2005, Pages 1515–1524, https://doi.org/10.1534/genetics.105.042366
Close - Share Icon Share
Abstract
Yeast Gtr1p and its human homolog RRAG A belong to the Ras-like small G-protein superfamily and genetically interact with RCC1, a guanine nucleotide exchange factor for Ran GTPase. Little is known regarding the function of Gtr1p. We performed yeast two-hybrid screening using Gtr1p as the bait to find interacting proteins. Rpc19p, a shared subunit of RNA polymerases I and III, associated with Gtr1p. The association of Gtr1p with Rpc19p occurred in a GTP-form-specific manner. RRAG A associated with RPA16 (human Rpc19p homolog) in a GTP-form-specific manner, suggesting that the association is conserved during evolution. Ribosomal RNA and tRNA synthesis were reduced in the gtr1Δ strain expressing the GDP form of Gtr1p, but not the GTP form of Gtr1p. Gel-filtration studies revealed an accumulation of the smaller Rpc19p-containing complex, but not of A135, in the gtr1Δ strain. Here, we propose that Gtr1p is involved in RNA polymerase I and III assembly by its association with Rpc19p and could be a mediator that links growth regulatory signals with ribosome biogenesis.
GUANINE nucleotide-binding proteins are a superfamily of regulatory GTP hydrolases composed of a large number of proteins, which include Ras family proteins, heterotrimeric G-protein α-subunits, and elongation factors TU and G, among others (Sprang 1997). They have crucial roles in cell growth, proliferation, differentiation, and macromolecular trafficking across different intracellular compartments (Milburn et al. 1990; Exton 1998). In yeast, Ras-like small G-proteins, including Ras1p, Ypt1p, Cdc42p, Arf1p, Gtr1p, and Gsp1p family proteins, bind to the guanine nucleotides GTP and GDP to function as molecular switches. Heterodimer formation of Gtr1p with Gtr2p is a feature that differs from other small G-proteins, which are monomeric.
Among the Ras superfamily of small G-proteins, Ran/Gsp1p is a nuclear protein with several functions, including nucleocytoplasmic transport of many types of protein and nucleic acids (Nishimoto 2000; Sazer and Dasso 2000). The guanine nucleotide exchange factor for Ran/Gsp1p (RCC1/Prp20p) is confined within the nucleus (Quimby et al. 2000; Nemergut et al. 2001; Li et al. 2003), whereas the Ran GTPase-activating protein (RanGAP/Rna1p) is located in the cytosol. Compartmentalization of these factors is believed to create a gradient of GTPase Ran across the nuclear pore complex, which controls the stability of importin-β interactions with particular cargo molecules. The Ran gradient is also a key factor that controls mitotic processes, including spindle assembly during metaphase and reformation of the nuclear envelope during telophase (Azuma and Dasso 2000; Heald and Weis 2000; Quimby et al. 2000).
A cold-sensitive mutant of GTR1, gtr1-11, was identified as a suppressor of mtr1-2, a temperature-sensitive mutant of the Saccharomyces cerevisiae RCC1 homolog and rna1-1, a temperature-sensitive mutant of Gsp1p GTPase-activating protein (Nakashima et al. 1996). Gtr1p genetically interacts with Pho84p, a phosphate transporter (Bun et al. 1992). Gtr1p forms complexes with itself and Gtr2p, a member of the Gtr1p subfamily of Ras-like small G-proteins, and negatively regulates the Ran/Gsp1p cycle through Gtr2p (Nakashima et al. 1999). RRAG A/Rag A is a functional human homolog of Gtr1p (Hirose et al. 1998) that interacts with RRAG C/Rag C and RRAG D/Rag D GTP-binding proteins (Sekiguchi et al. 2001), as well as NOP132 nucleolar protein (Sekiguchi et al. 2004). The yeast Nop8p is a Nip7p-interacting protein involved in 60S ribosome biogenesis that also interacts with Gtr1p (Ito et al. 2000; Sekiguchi et al. 2004). In Nop8p-depleted cells, pre-ribosomal RNA (rRNA) processing is abnormal (Zanchin and Goldfarb 1999). S. cerevisiae Nip7p is required for efficient 60S ribosome biogenesis and is conserved evolutionarily (Zanchin et al. 1997).
In the yeast S. cerevisiae, as in other eukaryotes, synthesis of rRNA transcripts accounts for 60% of the total transcriptional activity of rapidly growing yeast cells, which takes place in the nucleolus and is catalyzed by RNA polymerase I (pol I), which contains 14 distinct polypeptides (Buhler et al. 1976; Valenzuela et al. 1976; Carles et al. 1991). Five of the polypeptides, Rpb5p, Rpb6p, Rpb8p, Rpb10p, and Rpc10p, encoded by RPB5, RPB6, RPB8, RPB10, and RPC10, respectively, are common to all three nuclear RNA polymerases. Two other subunits, Rpc40p and Rpc19p, which are similar to bacterial α-subunits, are shared by pol I and pol III and are encoded by RPC40 and RPC19, respectively (Mann et al. 1987; Dequard-Chablat et al. 1991). Of the remaining seven pol I subunits, the two large ones, Rpa190p and Rpa135p (encoded by RPA190 and RPA135), have sequence homology with the two large subunits of pol II, pol III, and the β- and β′-subunits of bacterial RNA polymerase (Memet et al. 1988; Yano and Nomura 1991). Pol III synthesizes the precursors of 5S rRNA, the tRNAs, and a variety of other small nuclear and cytosolic RNAs and comprises 18 subunits, including Rpc40p and Rpc19p (Chedin et al. 1998). Rpc19p might have an ancestral gene of archaeal origin, whereas Rpc40p might have a bacterial origin (Lalo et al. 1993)
Using two-hybrid screening, we searched for a novel protein that interacts with Gtr1p and determined that Rpc19p was associated with Gtr1p in a GTP-form-dependent manner. Human RRAG A was also associated with RPA16/POLR1D, suggesting that Gtr1p functions in the nucleolus are conserved evolutionarily. Thus, we examined RNA pol I and III activity in gtr1Δ and observed that their activity was downregulated in gtr1Δ, suggesting that Gtr1p is required for RNA polymerase I and III function. Gel filtration of proteins from gtr1Δ resulted in an accumulation of the smaller Rpc19p-containing complex, suggesting that Gtr1p influences assembly of RNA polymerase I and III multi-subunit complexes.
MATERIALS AND METHODS
Strains, media, and two-hybrid assay:
NBW5 gtr1Δ (MATα gtr1-1Δ trp1-289 leu2-3,112 ade2 ura3-1,2 can1) and wild-type NBW5 (MATα trp1-289 leu2-3,112 his3-532 ade2 ura3-1,2 can1) strains were grown in YPD (2% glucose, 2% peptone, and 1% yeast extract). NBW5gtr1Δ strains harboring pEG-KT containing RPC19 (pQ19) or pEG-KT and Yeplac112 containing HA-tagged GTR1 (pL80) were grown in SD −Ura, −Trp (2% glucose, 0.67% yeast nitrogen base without amino acids, supplemented with all the essential amino acids except for uracil and tryptophan). Transformation of S. cerevisiae was performed using the lithium-acetate method with dimethyl sulfoxide (Hill et al. 1991). Yeast strains were maintained at either 14° or 16° for the nonpermissive temperature and at 26° for the permissive temperature.
Yeast two-hybrid assay (Chien et al. 1991) was performed using the S. cerevisiae Y190 strain (a gal4 gal80 his3 trp1 ade2 ura3 leu2 URA3::GAL1-lacZ LYS2::GAL1-HIS3 cyhr) to test protein interactions in vivo, as described previously (Sekiguchi et al. 2001). Protein interaction was tested by histidine-phototropic growth on SD −T, −L, −H +3 −AT plates (2% glucose, 0.67% yeast nitrogen base without amino acids, supplemented with all the essential amino acids except for tryptophan, leucine, and histidine in the presence of 30 mm 3-aminotriazol). The liquid β-galactosidase assay was performed as described previously (Sekiguchi et al. 2001). The β-galactosidase chromogenic filter assays were performed by transferring the yeast colonies onto nitrocellulose filters (Protran BA85; Schleicher & Schuell, Dassel, Germany). The yeast cells were partially lysed by submerging the filters in liquid nitrogen for 1 min. Filters were processed as described previously (Sekiguchi et al. 2004). Color, representing a positive signal, appeared within 30–60 min at 30° (Figures 1 and 2)
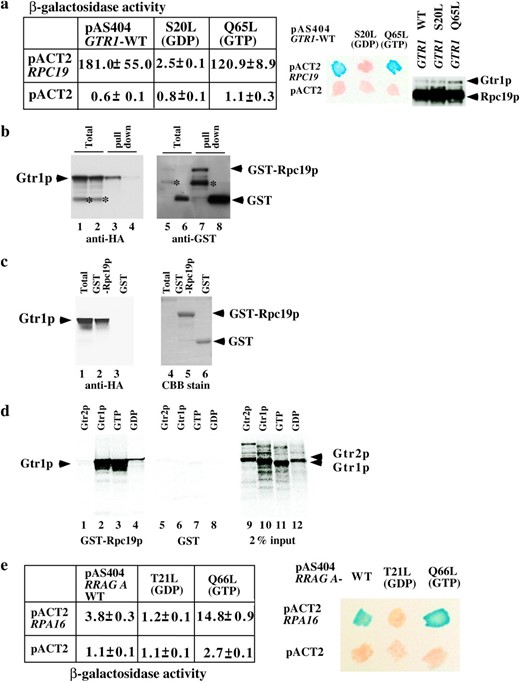
Association of Rpc19p with Gtr1p. (a) Extracts from cultures of S. cerevisiae colonies harboring two yeast genes, GTR1 wild type, S20L or the Q65L form in the pAS404 vector, or RPC19 in the pACT2 vector as shown, were obtained. Their β-galactosidase activities were measured as described before (Sekiguchi et al. 2001) and are shown as means of triplicate values with standard deviations. A β-galactosidase filter assay was also performed (middle). Patches of blue color appearing within 30 min represent a positive signal. White patches represent a negative signal. Expression of HA-tagged Gtr1p and Rpc19p in the yeast extracts described above was shown by immunoblotting against the HA tag (right). (b) Either GST-Rpc19p (lanes 1, 3, 5, and 7) or GST (lanes 2, 4, 6, and 8) was coexpressed with HA-tagged Gtr1p in NBW5 gtr1Δ. Prepared crude extracts were mixed with glutathione Sepharose-4B beads and pulled down as described previously (Nakashima et al. 1999). Bound proteins were run on SDS-polyacrylamide gels and transferred onto nylon membranes, which were immunoblotted with either anti-HA (lanes 1–4) or anti-GST (lanes 5–8) antibodies. Lanes 1, 2, 5, and 6: total proteins (10% input). An asterisk indicates degraded protein. (c) HA-tagged Gtr1p was expressed in NBW5 gtr1Δ. Yeast cell lysate was prepared and pulled down with either E. coli-produced GST-Rpc19p (lanes 2 and 5) or GST (lanes 3 and 6), which were bound to glutathione Sepharose-4B. Bound proteins were run on SDS-polyacrylamide gels and transferred onto nylon membranes, which were immunoblotted with the anti-HA (lanes 1–3) antibody. The filters were then stained with Coomassie Brilliant Blue R-250 (lanes 4–6). Lanes 1 and 4: total proteins (10% input). (d) 35S-labeled recombinant proteins (Gtr2p, lanes 1 and 5; Gtr1p, lanes 2 and 6; Gtr1p GTP form, lanes 3 and 7; the GDP form of Gtr1p, lanes 4 and 8) were produced with the TNT-Quick Coupled reaction kit and were pulled down with either GST-Rpc19p (lanes 1–4) or GST (lanes 5–8) protein, which was bound to glutathione Sepharose-4B beads. Bound proteins were run on SDS-polyacrylamide gels and analyzed using the Fuji image analyzer. Input proteins (2%) are shown in lanes 9–12. (e) Extracts from cultures of S. cerevisiae colonies harboring two human genes—RRAG A wild type (the T21L or the Q66L form in the pAS404 vector) and RPA16 in the pACT2 vector as shown—were obtained. Their β-galactosidase activities are shown as means of triplicate values with standard deviations. β-Galactosidase filter assay using Y190 strains harboring pAS404-RRAG A, pAS404-RRAG A(T21L) (GDP form), or pAS404-RRAG A(Q66L) (GTP form) and pACT2-RPA16 was performed as described above.
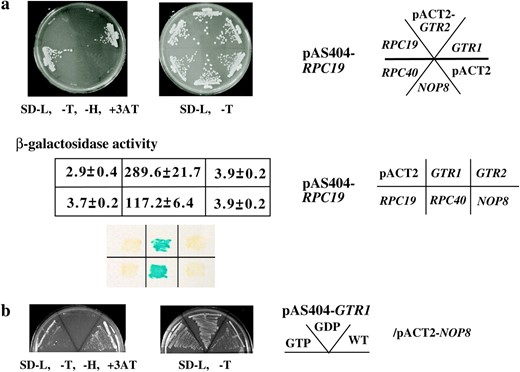
Mode of association of Gtr1p with Rpc19p and Nop8p. (a) Y190 strains harboring pAS404-RPC19 and pACT2-RPC19, pACT2-RPC40, pACT2-GTR1, pACT2-GTR2, pACT2-NOP8, or pACT2 vector were plated on SD −L, −T or SD −L, −T, −H, +3-AT plate at 30° for 2 or 5 days. Extracts from cultures of S. cerevisiae colonies harboring two yeast genes, RPC19 in the pAS404 vector and GTR1, GTR2, RPC19, RPC40, or NOP8 in the pACT2 vector as shown—were obtained. Their β-galactosidase activities are shown as means of triplicate values with standard deviations. A β-galactosidase filter assay was performed (bottom). (b) Y190 strains harboring pAS404-GTR1, pAS404-gtr1-11 (S20L) (GDP form), or pAS404-gtr1-13 (Q65L) (GTP form) and pACT2-NOP8 were plated on SD −L, −T or SD −L, −T, −H, +3-AT plate at 30° for 2 or 5 days.
.
Plasmid construction:
Plasmids used in this study
Name . | Yeast genetic marker . | Origin . | Source or reference . |
|---|---|---|---|
| pL20 | pGEX-KG containing GTR1 | Nakashima et al. (1999) | |
| pL21 | 2μ LEU2 GAL4AD GTR1 | GAL4 AD-fused GTR1 | Nakashima et al. (1999) |
| pL44 | TRP1GAL4DBD-GTR1 | GAL4DBD-fused GTR1 for two-hybrid assay | Nakashima et al. (1999) |
| pL66 | TRP1GAL4DBD-gtr1-11 | GAL4DBD-fused gtr1-11 for two-hybrid assay | Nakashima et al. (1999) |
| pL80 | 2μ TRP1 | YEplac112 containing HA-tagged GTR1 | Nakashima et al. (1999) |
| pL106 | TRP1GAL4DBD-gtr1Q65L | GAL4DBD-fused gtr1Q65L for two-hybrid assay | Nakashima et al. (1999) |
| pL115 | pET28a containing GTR1 | Nakashima et al. (1999) | |
| pL116 | pET28a containing gtr1-11 | This study | |
| pL118 | pET28a containing gtr1Q65L | Nakashima et al. (1999) | |
| pL242 | URA3 GTR1 | pRS406 containing GTR1 | This study |
| pL243 | URA3 gtr1-11 | pRS406 containing gtr1-11 allele | This study |
| pL244 | URA3 gtr1-Q65L | pRS406 containing gtr1-Q65L | This study |
| pL150 | 2μ LEU2 GAL4AD-GTR2 | GAL4 AD-fused GTR2 | Nakashima et al. (1999) |
| pL197 | pET28a containing GTR2 | This study | |
| pQ1 | TRP1 GAL4DBD-RRAG A | GAL4DBD-fused RRAG A for two-hybrid assay | Sekiguchi et al. (2001) |
| pQ2 | TRP1 GAL4DBD-RRAG A(T21L) | GAL4DBD-fused RRAG A(T21L) for two-hybrid assay | Sekiguchi et al. (2001) |
| pQ3 | TRP1 GALD4BD-RRAG A(Q65L) | GAL4DBD-fused RRAG A(Q65L) for two-hybrid assay | Sekiguchi et al. (2001) |
| pQ10 | 2μ LEU2 GAL4AD-RPC19 | GAL4 AD-fused RPC19 (AC19) | This study |
| pQ11 | TRP1 GAL4DBD-RPC19 | GAL4DBD-fused RPC19 for two-hybrid assay | This study |
| pQ12 | 2μ LEU2 GAL4AD-RPC40 | GAL4 AD-fused RPC40 (AC40) | This study |
| pQ13 | 2μ LEU2 GAL4AD-NOP8 | GAL4 AD-fused NOP8 | Sekiguchi et al. (2004) |
| pQ16 | 2μ URA3 GTR1 | YEplac195 containing RPC19 | This study |
| pQ17 | 2μ LEU2 GAL4AD-RPA16 | GAL4 AD-fused RPA16 | This study |
| pQ18 | pGEX-KG containing RPC19 (AC19) | This study | |
| pQ19 | 2μ URA3leu2-d GAL1-10:AC19 | pEG-KT containing RPC19 | This study |
Name . | Yeast genetic marker . | Origin . | Source or reference . |
|---|---|---|---|
| pL20 | pGEX-KG containing GTR1 | Nakashima et al. (1999) | |
| pL21 | 2μ LEU2 GAL4AD GTR1 | GAL4 AD-fused GTR1 | Nakashima et al. (1999) |
| pL44 | TRP1GAL4DBD-GTR1 | GAL4DBD-fused GTR1 for two-hybrid assay | Nakashima et al. (1999) |
| pL66 | TRP1GAL4DBD-gtr1-11 | GAL4DBD-fused gtr1-11 for two-hybrid assay | Nakashima et al. (1999) |
| pL80 | 2μ TRP1 | YEplac112 containing HA-tagged GTR1 | Nakashima et al. (1999) |
| pL106 | TRP1GAL4DBD-gtr1Q65L | GAL4DBD-fused gtr1Q65L for two-hybrid assay | Nakashima et al. (1999) |
| pL115 | pET28a containing GTR1 | Nakashima et al. (1999) | |
| pL116 | pET28a containing gtr1-11 | This study | |
| pL118 | pET28a containing gtr1Q65L | Nakashima et al. (1999) | |
| pL242 | URA3 GTR1 | pRS406 containing GTR1 | This study |
| pL243 | URA3 gtr1-11 | pRS406 containing gtr1-11 allele | This study |
| pL244 | URA3 gtr1-Q65L | pRS406 containing gtr1-Q65L | This study |
| pL150 | 2μ LEU2 GAL4AD-GTR2 | GAL4 AD-fused GTR2 | Nakashima et al. (1999) |
| pL197 | pET28a containing GTR2 | This study | |
| pQ1 | TRP1 GAL4DBD-RRAG A | GAL4DBD-fused RRAG A for two-hybrid assay | Sekiguchi et al. (2001) |
| pQ2 | TRP1 GAL4DBD-RRAG A(T21L) | GAL4DBD-fused RRAG A(T21L) for two-hybrid assay | Sekiguchi et al. (2001) |
| pQ3 | TRP1 GALD4BD-RRAG A(Q65L) | GAL4DBD-fused RRAG A(Q65L) for two-hybrid assay | Sekiguchi et al. (2001) |
| pQ10 | 2μ LEU2 GAL4AD-RPC19 | GAL4 AD-fused RPC19 (AC19) | This study |
| pQ11 | TRP1 GAL4DBD-RPC19 | GAL4DBD-fused RPC19 for two-hybrid assay | This study |
| pQ12 | 2μ LEU2 GAL4AD-RPC40 | GAL4 AD-fused RPC40 (AC40) | This study |
| pQ13 | 2μ LEU2 GAL4AD-NOP8 | GAL4 AD-fused NOP8 | Sekiguchi et al. (2004) |
| pQ16 | 2μ URA3 GTR1 | YEplac195 containing RPC19 | This study |
| pQ17 | 2μ LEU2 GAL4AD-RPA16 | GAL4 AD-fused RPA16 | This study |
| pQ18 | pGEX-KG containing RPC19 (AC19) | This study | |
| pQ19 | 2μ URA3leu2-d GAL1-10:AC19 | pEG-KT containing RPC19 | This study |
Plasmids used in this study
Name . | Yeast genetic marker . | Origin . | Source or reference . |
|---|---|---|---|
| pL20 | pGEX-KG containing GTR1 | Nakashima et al. (1999) | |
| pL21 | 2μ LEU2 GAL4AD GTR1 | GAL4 AD-fused GTR1 | Nakashima et al. (1999) |
| pL44 | TRP1GAL4DBD-GTR1 | GAL4DBD-fused GTR1 for two-hybrid assay | Nakashima et al. (1999) |
| pL66 | TRP1GAL4DBD-gtr1-11 | GAL4DBD-fused gtr1-11 for two-hybrid assay | Nakashima et al. (1999) |
| pL80 | 2μ TRP1 | YEplac112 containing HA-tagged GTR1 | Nakashima et al. (1999) |
| pL106 | TRP1GAL4DBD-gtr1Q65L | GAL4DBD-fused gtr1Q65L for two-hybrid assay | Nakashima et al. (1999) |
| pL115 | pET28a containing GTR1 | Nakashima et al. (1999) | |
| pL116 | pET28a containing gtr1-11 | This study | |
| pL118 | pET28a containing gtr1Q65L | Nakashima et al. (1999) | |
| pL242 | URA3 GTR1 | pRS406 containing GTR1 | This study |
| pL243 | URA3 gtr1-11 | pRS406 containing gtr1-11 allele | This study |
| pL244 | URA3 gtr1-Q65L | pRS406 containing gtr1-Q65L | This study |
| pL150 | 2μ LEU2 GAL4AD-GTR2 | GAL4 AD-fused GTR2 | Nakashima et al. (1999) |
| pL197 | pET28a containing GTR2 | This study | |
| pQ1 | TRP1 GAL4DBD-RRAG A | GAL4DBD-fused RRAG A for two-hybrid assay | Sekiguchi et al. (2001) |
| pQ2 | TRP1 GAL4DBD-RRAG A(T21L) | GAL4DBD-fused RRAG A(T21L) for two-hybrid assay | Sekiguchi et al. (2001) |
| pQ3 | TRP1 GALD4BD-RRAG A(Q65L) | GAL4DBD-fused RRAG A(Q65L) for two-hybrid assay | Sekiguchi et al. (2001) |
| pQ10 | 2μ LEU2 GAL4AD-RPC19 | GAL4 AD-fused RPC19 (AC19) | This study |
| pQ11 | TRP1 GAL4DBD-RPC19 | GAL4DBD-fused RPC19 for two-hybrid assay | This study |
| pQ12 | 2μ LEU2 GAL4AD-RPC40 | GAL4 AD-fused RPC40 (AC40) | This study |
| pQ13 | 2μ LEU2 GAL4AD-NOP8 | GAL4 AD-fused NOP8 | Sekiguchi et al. (2004) |
| pQ16 | 2μ URA3 GTR1 | YEplac195 containing RPC19 | This study |
| pQ17 | 2μ LEU2 GAL4AD-RPA16 | GAL4 AD-fused RPA16 | This study |
| pQ18 | pGEX-KG containing RPC19 (AC19) | This study | |
| pQ19 | 2μ URA3leu2-d GAL1-10:AC19 | pEG-KT containing RPC19 | This study |
Name . | Yeast genetic marker . | Origin . | Source or reference . |
|---|---|---|---|
| pL20 | pGEX-KG containing GTR1 | Nakashima et al. (1999) | |
| pL21 | 2μ LEU2 GAL4AD GTR1 | GAL4 AD-fused GTR1 | Nakashima et al. (1999) |
| pL44 | TRP1GAL4DBD-GTR1 | GAL4DBD-fused GTR1 for two-hybrid assay | Nakashima et al. (1999) |
| pL66 | TRP1GAL4DBD-gtr1-11 | GAL4DBD-fused gtr1-11 for two-hybrid assay | Nakashima et al. (1999) |
| pL80 | 2μ TRP1 | YEplac112 containing HA-tagged GTR1 | Nakashima et al. (1999) |
| pL106 | TRP1GAL4DBD-gtr1Q65L | GAL4DBD-fused gtr1Q65L for two-hybrid assay | Nakashima et al. (1999) |
| pL115 | pET28a containing GTR1 | Nakashima et al. (1999) | |
| pL116 | pET28a containing gtr1-11 | This study | |
| pL118 | pET28a containing gtr1Q65L | Nakashima et al. (1999) | |
| pL242 | URA3 GTR1 | pRS406 containing GTR1 | This study |
| pL243 | URA3 gtr1-11 | pRS406 containing gtr1-11 allele | This study |
| pL244 | URA3 gtr1-Q65L | pRS406 containing gtr1-Q65L | This study |
| pL150 | 2μ LEU2 GAL4AD-GTR2 | GAL4 AD-fused GTR2 | Nakashima et al. (1999) |
| pL197 | pET28a containing GTR2 | This study | |
| pQ1 | TRP1 GAL4DBD-RRAG A | GAL4DBD-fused RRAG A for two-hybrid assay | Sekiguchi et al. (2001) |
| pQ2 | TRP1 GAL4DBD-RRAG A(T21L) | GAL4DBD-fused RRAG A(T21L) for two-hybrid assay | Sekiguchi et al. (2001) |
| pQ3 | TRP1 GALD4BD-RRAG A(Q65L) | GAL4DBD-fused RRAG A(Q65L) for two-hybrid assay | Sekiguchi et al. (2001) |
| pQ10 | 2μ LEU2 GAL4AD-RPC19 | GAL4 AD-fused RPC19 (AC19) | This study |
| pQ11 | TRP1 GAL4DBD-RPC19 | GAL4DBD-fused RPC19 for two-hybrid assay | This study |
| pQ12 | 2μ LEU2 GAL4AD-RPC40 | GAL4 AD-fused RPC40 (AC40) | This study |
| pQ13 | 2μ LEU2 GAL4AD-NOP8 | GAL4 AD-fused NOP8 | Sekiguchi et al. (2004) |
| pQ16 | 2μ URA3 GTR1 | YEplac195 containing RPC19 | This study |
| pQ17 | 2μ LEU2 GAL4AD-RPA16 | GAL4 AD-fused RPA16 | This study |
| pQ18 | pGEX-KG containing RPC19 (AC19) | This study | |
| pQ19 | 2μ URA3leu2-d GAL1-10:AC19 | pEG-KT containing RPC19 | This study |
lists the plasmids used in this study. RPC19 was amplified with yeast total DNA using BamHI/KpnI site-tagged primers (RPC19N and RPC19C) and inserted into the BamHI/KpnI site of pEG-KT, resulting in pEG-KT-RPC19. The RPC19N and RPC19C sequences are 5′-GGGGATCCACATAACTTGCTTCTATTTTGGGA-3′ and 5′-GGGGTACCGAATACGCCTTTAAAAAGGAA-3′, respectively. Each construct was sequenced using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Purification of glutathione S-transferase-fused proteins:
Escherichia coli BL21(DE3) transformed with pGEX-KG-RPC19, pGEX-KG-GTR1, or pGEX-KG vector was grown in 750 ml of Luria Bertani medium. The culture was induced with 0.2 mm isopropyl β-thiogalactoside and grown at 26° for 4 hr. The cells were collected by centrifugation and resuspended in 30 ml of the lysis buffer [50 mm Tris (pH 7.5), 150 mm NaCl, 2.5 mm MgCl2, 10% glycerol, 0.5% NP-40, 1 mm phenylmethylsulfonyl fluoride, 0.1 μg/ml aprotinin, and 1 mm dithiothreitol] and sonicated three times for 5 min on ice (Sonicator, Heat System-Ultrasonics, Plainview, NY), with a microtip (40% cycle and output set to 4). The lysate was centrifuged at 10,000 × g for 30 min at 4°. The supernatant was incubated in 1 ml of 50% slurry (v/v) glutathione-Sepharose 4B (Amersham Biosciences, Piscataway, NJ) for 60 min at 4° while rotating. The glutathione S-transferase (GST) beads were washed three times with 5 ml of lysis buffer.
In vitro-binding assay:
Gtr1p and other proteins were synthesized in vitro as follows: pET28a-GTR1 (pL115) was incubated with 20 μCi of [35S]methionine (NEN Life Science Products, Boston) and 40 μl Quick Master Mix of TNT T7 Quick Coupled Transcription/Translation System (Promega, Madison, WI) for 90 min at 30° following the manufacturer's guidelines. The resultant extract was diluted to 600 μl with the binding assay buffer [50 mm Tris (pH 8.0), 10 mm MgCl2, 0.1 mm GTP, 1 mg/ml bovine serum albumin, 1 mm dithiothreitol]. GST (20 μg) and GST-Rpc19p (20 μg), which were bound to the glutathione Sepharose-4B beads, were each mixed with 200 μl of extract and incubated for 60 min at 4° while rotating. The beads were washed five times with 500 μl of the binding assay buffer and suspended with 30 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. All the bound proteins in 30 μl SDS-PAGE sample buffer were run on SDS-polyacrylamide gels and analyzed by a Fuji BAS2500 Image Analyzer (Fuji Photo Film, Tokyo).
Labeling of RNAs and Northern analysis:
Yeast cells were pulse labeled with 100 μCi/ml of methyl[3H]methionine for 2 min and chased with 100 μg/ml of unlabeled methionine. At various times, samples were taken and immediately stored at −80°. Total RNA was isolated from yeast cells using the hot-phenol method (Kohrer and Domdey 1991). Small RNA molecules were analyzed using 10% polyacrylamide gels containing 8.3 m urea in 1× TBE buffer. RNAs were then electrophoretically transferred to Hybond nylon membranes (Amersham Biosciences) in 0.5× TBE buffer. Large RNA molecules were separated by electrophoresis on 1.2% agarose-6% formaldehyde gels in 1× MOPS buffer and transferred to Hybond nylon membranes by capillary transfer using 20× SSC. For Northern analysis, after transfer, the membranes were dried for 10 min and the RNA was crosslinked by UV irradiation. Prehybridization and hybridization were performed overnight in 6× SSC, 0.1% SDS, 2× Denhardt's, and 125 μg/ml denatured salmon sperm DNA at 40°. The oligonucleotide probe 5′-GTGCATTTCGATTTGAAA-3′ was used to detect 5′ leader-containing pre-tRNA Leu3(CAA, 132 nt). U4 RNA (160 nt) was detected using the probe 5′-CCATGAGGAGACGGTCTGG-3′. The oligonucleotides (20 pmol) were end-labeled in 40-μl reactions using [γ-32P]ATP 50 μCi, 3000 Ci/mmol (Amersham Biosciences), and T4 polynucleotide kinase. Hybridization was detected using the imaging plate and Fuji image analyzer (Fuji Photo film). RNA levels were normalized using the signal for U4 RNA.
Immunoblotting and antibodies:
Protein samples were electrophoresed on a 10 or 12.5% SDS-polyacrylamide slab gel or 5–20% gradient gel (PAGEL, Atto, Tokyo, Japan) and analyzed by immunoblotting. Immunoblotting was performed as described previously (Sekiguchi et al. 2001) using an ECL kit (Amersham Biosciences) as recommended by the supplier. Mouse antihemagglutinin (anti-HA) antibody (catalog no. MMS-101P) was purchased from CRP (Cumberland, VA). The anti-Rpc19p antibody was raised against GST-Rpc19p antibody. The anti-A135 antibody was a gift from M. Nomura (University of California, Irvine, CA) and the anti-pol I antibody was a gift from M. Riva (Service de Biochimie et de Genetique Moleculaire, France).
Gel filtration:
Yeast proteins were size fractionated following the procedure described previously (Ebbert et al. 1999; Wang et al. 2004). Strain NBW5 gtr1Δ (containing the HA-GTR1-fused gene) or strain NBW gtr1Δ was cultivated in YPD medium at 30° and harvested at an OD600 of ∼1.0. Cells were resuspended in GF buffer (40 mm Hepes, pH 7.5; 120 mm NaCl; 0.1% Tween 20; 10% glycerol; 1 mm phenylmethylsulfonyl fluoride) and lysed by intensive shaking in the presence of an equal volume of glass beads for 1 min 10 times with 2-min intervals. The crude supernatant was clarified by centrifugation at 100,000 × g for 30 min. A Sephacryl S-300 HR column (Amersham Biosciences) was used for gel filtration. Fractions (5 ml) were collected and separated by gradient SDS-PAGE (5–20%).
RESULTS
Interaction between Gtr1p and Rpc19p depends on the bound nucleotide state:
A two-hybrid screening method was employed to identify interacting proteins using Gtr1p as bait. Rpc19p, which is a shared subunit of RNA pol I and III, was identified as a protein that interacts with Gtr1p (Figure 1a). Uetz et al. 2000 consistently obtained the same result using large-scale yeast two-hybrid screens. In general, Ras-like small G-proteins function as a binary switch by binding to GDP or GTP. Therefore, we examined whether the association of Gtr1p with Rpc19p depended on the guanine nucleotide binding state of Gtr1p. A yeast two-hybrid assay using a S20L mutant (GDP form) and Q65L mutant (GTP form) of Gtr1p (Nakashima et al. 1999) was performed. Y190 strains that harbor pACT2-RPC19 and either pAS404-GTR1 or pAS404-gtr1Q65L (GTP form) had 181 and 120.9 β-galactosidase units, respectively (Figure 1a, left), while Y190 strains that harbor pACT2-RPC19 and pAS404-gtr1S20L had 2.5 β-galactosidase units. Y190 strains that harbor control pairs had <1.1 β-galactosidase units. Yeast two-hybrid β-galactosidase filter assay was also conducted (Figure 1a, middle). These results indicate that Rpc19p might be an effector of Gtr1p and that its activity is influenced by a GTP-bound form of Gtr1p. We examined whether the Gal4 DNA-bound region-Gtr1p fusion proteins were expressed in those strains (Figure 1a, right). Rpc19p is associated with Rpc40p in the pol I and III complexes (Lalo et al. 1993). Thus, we examined whether the interaction between Rpc19p and Gtr1p occurred directly or indirectly. The interaction between Rpc40p and Gtr1p was not detected in the yeast two-hybrid assay (data not shown), indicating that the interaction between Rpc19p and Gtr1p was not mediated by Rpc40p. To further confirm the association of Gtr1p with Rpc19p in vivo, either GST fused to Rpc19p (GST-Rpc19p) or GST was coexpressed with HA-tagged Gtr1p in the gtr1Δ strain. GST-Rpc19p pulled down Gtr1p efficiently (Figure 1b, lane 3). As a control, GST protein failed to pull down Gtr1p (Figure 1b, lane 4). E. coli-produced GST-Rpc19p also pulled down HA-tagged Gtr1p expressed in the gtr1Δ strain (Figure 1c, lane 2). E. coli-produced GST failed to pull down Gtr1p (Figure 1c, lane 3). To confirm the guanine nucleotide-dependent association of Gtr1p with Rpc19p, GST-Rpc19p was used to pull down in vitro-synthesized Gtr1p or the GTP form of Gtr1p proteins (Figure 1d). The in vitro-synthesized Gtr1p and the GTP form of Gtr1p were efficiently pulled down by GST-Rpc19p (Figure 1d, lanes 2 and 3), while the GDP form of Gtr1p was not pulled down efficiently (Figure 1d, lane 4). Gtr2p was not pulled down by GST-Rpc19p at all (Figure 1d, lane 1). GST protein used as a control failed to pull down Gtr1p or Gtr2p (Figure 1d, lanes 5–8).
Amino acid sequence comparison revealed that RPA16/POLR1D is a human homolog of Rpc19p, (Hu et al. 2002). Human RRAG A is interchangeable with yeast Gtr1p. Thus, we examined whether RPA16 associated with RRAG A. Yeast two-hybrid β-galactosidase liquid and filter assays were conducted as described above (Figure 1e). Y190 strains harboring the wild-type and the GTP form of pAS404-RRAG A and pACT2-RPA16 had 3.8 and 14.8 β-galactosidase units, respectively, while those harboring the GDP form of pAS404-RRAG A and pACT2 had 1.2 β-galactosidase units. These findings suggest that RRAG A interacts with RPA16 in a GTP-form-specific manner, although the β-galactosidase activity of the RRAG A and RPA16 pairs is weak when compared with that of Gtr1p and Rpc19p. Y190 strains harboring RRAG A and a control pACT2 vector had 1.1–2.7 β-galactosidase units. These results indicate that the interaction between Gtr1p and Rpc19p is evolutionarily well conserved.
Association of Rpc19p with Gtr1p was observed as well as the association of Rpc19p with Rpc40p:
Rpc19p and Rpc40p form a complex to function as basic components of RNA pol I and III, as described in the Introduction. The β-galactosidase assay indicated that the Rpc19p associates with Gtr1p. Y190 strains harboring pAS404-RPC19 and pACT2-RPC40 had 117.2 β-galactosidase units, while those harboring pAS404-RPC19 and pACT2-GTR1 had 289.6 β-galactosidase units. Thus, the association between Gtr1p and Rpc19p was observed as well as the association between Rpc19p with Rpc40p (Figure 2a, middle). Control pairs had fewer than four β-galactosidase units. An association between Rpc19p with Gtr2p, Nop8p, or Rpc19p was not detected.
Gtr1p also associates with Nop8p. To confirm the previous results and examine the nucleotide-specific interactions of Gtr1p with its associating proteins, two-hybrid assays were performed (Figure 2, a and b). The association of Gtr1p with Nop8p also depended on the nucleotide bound state as shown by the plate assay (Figure 2b). The β-galactosidase activity of strains that harbor the pAS404-GTR1WT and pACT2-NOP8 pairs was at the same level as that of the controls (data not shown), although we previously demonstrated that Gtr1p efficiently pulled down Nop8p in vitro (Sekiguchi et al. 2004).
Ribosomal RNA synthesis was decreased in gtr1Δ:
gtr1Δ and gtr1-11 mutations (S20L) of Gtr1p exhibit a cold-sensitive phenotype at 14° on plates (Nakashima et al. 1996). To confirm that the cold sensitivity was also observed in liquid culture, the growth rate of wild-type, gtr1Δ, gtr1Δ (HA-tagged GTR1 transformed), gtr1Δ (HA-tagged GTR1 GDP form transformed), and gtr1Δ (HA-tagged GTR1 GTP form transformed) strains in YPD medium was measured at 26° (Figure 3a
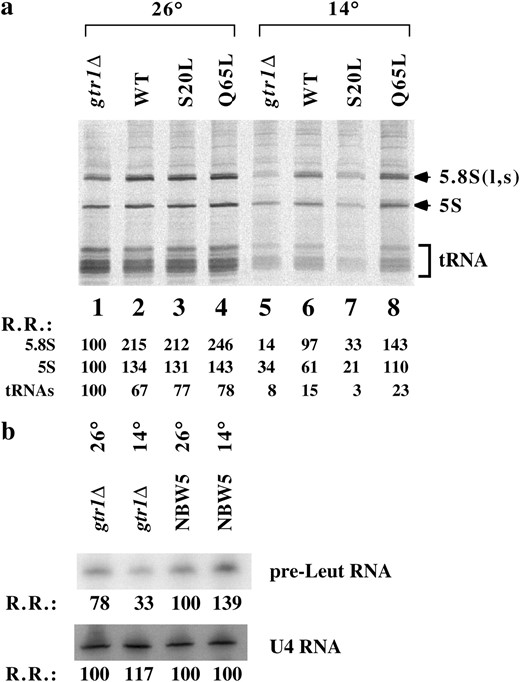
Ribosomal RNA synthesis was decreased in the gtr1Δ strain. (a) The growth curve of NBW5 gtr1Δ (•), NBW5 gtr1Δ expressing wild-type Gtr1p (pL242) (○), NBW5 gtr1Δ expressing Q65L Gtr1p (pL244) (□), NBW5 gtr1Δ expressing S20L Gtr1p (pL243) (▵), and wild-type NBW5 (+) strains at 26° (left) and at 14° (right) was studied as previously described (Zanchin et al. 1997). Decrease in the growth rate of gtr1Δ and S20L strains was significant at 14°. (b) To examine the global rRNA synthesis rate in the strains described above, exponentially growing cultures were labeled with [3H]uracil (50 μCi/ml) for 10 min at 26° or 30 min at 14° and cold excess uracil (300 μg/ml) was added and incubated to complete the processing for 1 hr at 26° or 3 hr at 14°. Extracted RNAs were run on formaldehyde agarose gel and transferred to Hybond filters. The filters were exposed to an imaging plate (TR2040, Fuji Photo film). Lanes 1 and 5, NBW5 gtr1Δ; lanes 2 and 6, NBW5 gtr1Δ (pL242); lanes 3 and 7, NBW5 gtr1Δ (pL243); lanes 4 and 8, NBW5 gtr1Δ (pL244). Relative radioactivity (R.R.) of 25S or 18S rRNA is shown at the bottom left (gtr1Δ, 26° = 100). RNA was stained with ethidium bromide showing that an equal amount of RNA was loaded on the gel (right). (c) Cells grown at 30° to midlog phase. 5′ ITS1 rRNA was localized by fluorescence in situ hybridization. Chromosomal DNA was labeled with 4′,6-diamidino-2-phenylindole-dihydrochloride to identify the location of the nucleus. (d) Total RNA was isolated from logarithmically grown gtr1Δ, gtr2Δ, and wild-type strains at either 26° or 16° for 2 hr. Total RNA (5 μg) was run on formaldehyde agarose gels and transferred onto nylon membranes and blotted with the 32P-labeled probes indicated at the right. Relative radioactivity (R.R.) of RPS3, RPL3, or ACT1 is shown at the bottom (wild type, 26° = 100). (e) rRNA processing was examined by 2-min pulse labeling with [3H]methyl methionine and chased for 2, 6, and 15 min in the presence of methionine (100 μg/ml), as described previously (Zanchin et al. 1997). Positions of 27S, 25S, 20S, and 18S are indicated.
, left) and 14° (Figure 3a, right). Growth retardation of the gtr1Δ and gtr1Δ (GDP form) strains was observed at 14° and at 26° and rescued by the expression of wild type or GTP-form Gtr1p. Ribosomal RNA synthesis was reduced in gtr1Δ and gtr1Δ (GDP form) at 14° (Figure 3b, left, lanes 5 and 7). Introduction of the wild-type GTR1 and GTP-form GTR1 into gtr1Δ recovered the impaired RNA synthesis rate (Figure 3b, left, lanes 6 and 8). rRNA synthesis occurred in all strains at 26° (Figure 3b, left, lanes 1–4). Ethidium bromide staining of RNA indicated that a similar amount of RNA was run on the agarose gel (Figure 3b, right). Consistently, in situ hybridization of the rRNA precursor with the 5′ internal transcribed spacer (ITS) probe (Moy and Silver 2002) revealed that there were smaller amounts of the rRNA precursor in gtr1Δ than in wild-type cells (Figure 3c), suggesting that the rRNA synthesis rate in each cell was decreased in the gtr1Δ strain.
In yeast, a decrease in rRNA synthesis accompanies a decrease in ribosomal protein synthesis (i.e., the stringent response) (Warner 1999). Thus, to examine whether ribosomal protein mRNA expression was decreased in the gtr1Δ strain, we conducted Northern blot analysis using RNA prepared from wild-type, gtr1Δ, and gtr2Δ strains at 26° and at 16° for 4 hr. gtr1Δ strains exhibited cold sensitivity at both 14° and 16°. The expression of Rps3 and Rpl3 ribosomal protein mRNAs was decreased in gtr1Δ, but not in gtr2Δ, when compared with those in the wild-type strain (Figure 3d). The RNA content of each sample of Northern blotting was the same as that demonstrated by ethidium bromide staining (Figure 3d, bottom). In support of this finding, the expression of many ribosomal protein mRNAs decreased markedly in gtr1Δ in the preliminary microarray experiment (data not shown).
Gtr1p is also associated with Nop8p, which is involved in rRNA processing. Thus, we examined rRNA processing in both the wild-type and Gtr1p-disrupted strains using conventional pulse-chase experiments with [3H]methionine at 26°. Processing of 27S rRNA occurred rapidly in the wild-type strain (within 3 min; Figure 3e). In the Gtr1p-disrupted strain, rRNA processing of 27S rRNA required more time than did wild-type cells (within 6 min). Similar results were obtained at 14° (data not shown). The processing of 18S rRNA in the Gtr1p-disrupted strain also required more time when compared with that in the wild-type strain, indicating that Gtr1p is not essential for rRNA processing, but that it might accelerate the rRNA processing rate. The polysome profile of wild-type and gtr1Δ strains was analyzed at both 26° and 16°. There was no obvious change in the ratio between 60S and 40S in either the wild-type or the gtr1Δ strains, however, except a reduced 80S monosome level in the gtr1Δ strain, which might be due to reduced rRNA synthesis in the gtr1Δ strain (data not shown).
Decreased RNA pol III activity in the gtr1Δ strain:
Rpc19p is a subunit shared between RNA pol I and III. We asked whether RNA pol III activity was also decreased in the gtr1Δ strain at a nonpermissive temperature (Figure 4)
RNA pol III activity was also decreased in gtr1Δ strain. (a) Equal amounts of RNA samples from Figure 3b were separated with 10% polyacrylamide gels and transferred onto Hybond filters. Radioactivity was detected using a Fuji image analyzer. Relative radioactivity (R.R.) is shown at the bottom with the count of gtr1Δ at 26° assumed to be 100. This experiment was performed twice with similar results. (b) Northern blot analysis of precursor RNAs transcribed by RNA pol III. Total RNA from wild-type and gtr1Δ strains was analyzed for pr-tRNALeu3 or U4 RNA as indicated. R.R. is shown (NBW5, 26° = 100)
. Pulse labeling of RNA with [3H]uracil revealed that the 5S and tRNA synthesis rates were reduced in gtr1Δ expressing the GDP form of Gtr1p at 14° (Figure 4a, lane 7). Introduction of wild-type and GTP-form GTR1 into gtr1Δ recovered the impaired RNA synthesis rate (Figure 4a, lanes 6 and 8). It should be noted that 5.8S synthesis reduction was greater compared with 5S and tRNA synthesis reduction in the gtr1Δ strain (Figure 4a, lane 5), whereas the small RNA synthesis in gtr1Δ expressing the GDP form of Gtr1p was markedly reduced (Figure 4a, lane 7), suggesting that the gtr1Δ strain has properties slightly different from those of the gtr1Δ strain expressing the GDP form of Gtr1p. We used another method to examine RNA pol III activity in gtr1Δ (Figure 4b). A reduction in pre-Leu tRNA synthesis was also observed, suggesting that RNA pol I and III activities were decreased in the gtr1Δ strain at a nonpermissive temperature.
Accumulation of the smaller Rpc19p-containing complex in the gtr1Δ strain:
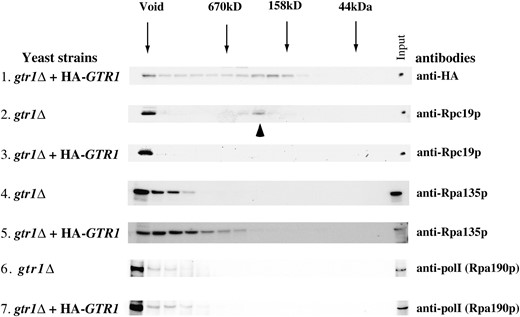
Finally, we examined how Gtr1p influences Rpc19p function. Gel-filtration analysis indicated that Gtr1p was present in the void fraction and in the lower fractions (Figure 5
Gel-filtration analysis of Gtr1p in wild-type and gtr1Δ strains. Gtr1p appears at the same location (void) with Rpc19p in addition to the smaller molecular weight fractions in the S300 gel-filtration analysis. Disruption of GTR1 leads to an accumulation of the smaller Rpc19p-containing complex. Accumulation of A135 and A190 in the same fraction as Rpc19p was not observed in the gtr1Δ strain. (Top) HA-tagged Gtr1p was detected with anti-HA antibody. Molecular mass is indicated above by arrows. Yeast strains are indicated at the left: gtr1Δ, NBW5gtr1Δ; gtr1Δ + HA-GTR1, NBW5 gtr1Δ expressing HA-tagged Gtr1p. This experiment was performed twice with similar results.
, top). A significant amount of Rpc19p was present in the smaller-size columns (indicated by an arrowhead) as well as in the void fraction in the gtr1Δ strain (Figure 5, second panel). Introduction of HA-tagged GTR1 into the gtr1Δ strain resulted in the localization of Rpc19p in the void fraction of >670 kDa (Figure 5, third panel). To examine whether an association of the Rpc19p-containing complex with other pol I subunits becomes unstable in the absence of Gtr1p, a reblot with an antibody against A135 (anti-A135) and A190 (anti-pol I) was conducted and revealed no accumulation of A135- or A190-containing complexes in the gtr1Δ strain, indicating that the smaller-size Rpc19p fraction did not contain A135. Thus, in the absence of Gtr1p, the association of Rpc19p with an A135-containing large complex might be inefficient.
DISCUSSION
Negative regulation of the Ran/Gsp1p cycle by Gtr1p is an important issue (Nakashima et al. 1996, 1999). While much attention has focused on the function of Ran/Gsp1p, little attention has been paid to Gtr1p/RRAG A. We identified proteins associating with RRAG A and previously identified a novel nucleolar protein, NOP132, with a two-hybrid assay using RRAG A as bait (Sekiguchi et al. 2004). Here, we established the RNA polymerase subunit Rpc19p as a Gtr1p-associating protein using a two-hybrid assay with GTR1 as bait. The mode of association of Gtr1p with Rpc19p is GTP specific, suggesting that Rpc19p is an effector of Gtr1p.
Previously, we demonstrated that the yeast Nop8p, which is similar to NOP132, also interacts with Gtr1p (Sekiguchi et al. 2004). Here, we demonstrate that RPA16, which is similar to Rpc19p, interacts with RRAG A, suggesting that RPA16 has a function similar to Rpc19p in humans. Because Gtr1p and RRAG A proteins are evolutionarily well conserved (Hirose et al. 1998), it is likely that their target proteins are also well conserved. Thus, the Gtr1p/RRAG A system should be well conserved through evolution, if the Gtr1p and RRAG A proteins are involved in RNA polymerase activity, a basic mechanism for protein synthesis and essential for life. The fact that both Nop8p/NOP132 and Rpc19p/RPA16 proteins localize in the nucleolus to perform their roles in ribosome biogenesis suggests that Gtr1p/RRAG A has multiple functions in ribosome biogenesis in the nucleolus. In yeast, as in E. coli, there was a stringent response: coupling of ribosome RNA and protein synthesis. When yeast needs to grow slowly due to damage or a change in circumstances, ribosome biogenesis should simultaneously decelerate for some period of time at multiple points, such as during rRNA transcription, rRNA processing, etc., to save energy. Gtr1p might be this type of regulator, because it acts on at least two stages during ribosome biogenesis.
RNA pol I and III activity was reduced in gtr1 mutant strains:
Ribosomal RNA synthesis was reduced in the gtr1Δ strain at 14°, suggesting that the Gtr1p interaction with Rpc19p is important in rRNA synthesis by RNA pol I. Because both rRNA and ribosomal protein synthesis consume a lot of energy, ribosomal RNA synthesis is strictly coupled to ribosomal protein synthesis for the economy of the cell (Warner 1999). Here, we observed that expression of some ribosomal protein mRNAs was reduced at a low temperature in gtr1Δ, although we cannot rule out the possibility that the deletion of Gtr1p caused a stringent response and thus might be acting on factors upstream of RNA pol I. In gtr2Δ, such a decrease in the expression of ribosomal protein mRNAs was not observed (Figure 5 and data not shown), suggesting that the function of Gtr1p differs from that of Gtr2p. 5S and tRNA synthesis was decreased in the gtr1Δ strain and the gtr1Δ strain expressing the S20L mutation of Gtr1p at 14°, suggesting that Gtr1p is also involved in RNA pol III activity. These data are consistent with the fact that Rpc19p is a shared subunit of RNA pol I and III.
Accumulation of the smaller Rpc19p protein-containing complex:
The study of ribosome assembly in bacteria was accelerated using the large number of ribosome assembly mutants. It was found that a large percentage of cold-sensitive mutants have conditional blocks in ribosomal subunit assembly (Guthrie et al. 1969; Tai et al. 1969). In cold-sensitive mutants, nonstructural components as well as structural ribosomal protein are involved in ribosome assembly (Bryant and Sypherd 1974). We thus speculate that cold sensitivity of the gtr1 mutant results from assembly defects in RNA polymerase I and III multi-subunit complexes. Consistently, gel-filtration data suggest that accumulation of the smaller Rpc19p protein-containing complex increased in the absence of Gtr1p, suggesting that Gtr1p is involved in the complex formation of Rpc19p with other RNA polymerase subunits for rRNA transcription. It was previously proposed that Rpc40p, but not Rpc19p, is involved subunit assembly (Dequard-Chablat et al. 1991). Thus, Rpc19p might be involved in higher-order complex formation, such as that of RNA polymerase with initiation factors. Because a small fraction of Rpc19p (∼400 kDa) (Figure 5) did not contain A135, it was not composed of subunits (Rpc19p, Rpc40p, A190, A135) corresponding to the bacterial α2ββ′ subunit composition of the bacterial core enzyme, which is sufficient to form a functional enzyme. Gtr1p might have a role in the yeast pol I core subunit assembly between the Rpc19p-containing complex and the A135-containing complex by directly acting on Rpc19p to influence cell proliferation. While detailed molecular mechanisms underlying the change in the components of these complexes require further study, Gtr1 regulation in ribosome biogenesis could be a novel mechanism for cell growth and proliferation in Tor or other pathways.
When we examined the genetic interaction between GTR1 and RPC19, we did not observe suppression of gtr1Δ by RPC19 overexpression (data not shown). Because Gtr1p is a member of Ras-like small G-proteins, it might have multiple effectors to exert multiple functions like Ran, which might make it difficult to suppress GTR1 mutation by overexpression of RPC19. Global identification by mass spectrometry analysis will help identify more proteins that interact with Gtr1p. RRAG A and Gtr1p might function via their effectors, such as NOP132, Nop8p, RPA16, and Rpc19p, and still-unknown targets. Further biochemical analysis will clarify the functions of Gtr1p/RRAG A.
Footnotes
Communicating editor: M. Hampsey
Acknowledgement
We thank M. Nomura and M. Riva for providing us antibodies to A135 and pol I, respectively. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (T.S.) and by Grants-in-Aid for Specially Promoted Research (T.N.) from the Japan Ministry of Education, Science, Sport and Culture.
References
Azuma, Y., and M. Dasso,
Bryant, R. E., and P. S. Sypherd,
Buhler, J. M., F. Iborra, A. Sentenac and P. Fromageot,
Bun, Y. M., S. Harashima and Y. Oshima,
Carles, C., I. Treich, F. Bouet, M. Riva and A. Sentenac,
Chedin, S., M. L. Ferri, G. Peyroche, J. C. Andrau, S. Jourdain et al.,
Chien, C. T., P. L. Bartel, R. Sternglanz and S. Fields,
Dequard-Chablat, M., M. Riva, C. Carles and A. Sentenac,
Ebbert, R., A. Birkmann and H. J. Schuller,
Guthrie, C., H. Nashimoto and M. Nomura,
Heald, I., and I. Weis,
Hill, J., K. A. Donald, D. E. Griffiths and G. Donald,
Hirose, E., N. Nakashima, T. Sekiguchi and T. Nishimoto,
Hu, P., S. Wu, Y. Sun, C. C. Yuan, R. Kobayashi et al.,
Ito, T., K. Tashiro, S. Muta, R. Ozawa, T. Chiba et al.,
Kohrer, K., and H. Domdey,
Lalo, D., C. Carles, A. Sentenac and P. Thuriaux,
Li, H. Y., D. Wirtz and Y. Zheng,
Mann, C., J. M. Buhler, I. Treich and A. Sentenac,
Memet, S., M. Gouy, C. Marck, A. Sentenac and J. M. Buhler,
Milburn, M. V., L. Tong, A. M. DeVos, A. Brunger, Z. Yamaizumi et al.,
Moy, T. I., and P. A. Silver,
Nakashima, N., N. Hayashi, E. Noguchi and T. Nishimoto,
Nakashima, N., E. Noguchi and T. Nishimoto,
Nemergut, M. E., C. A. Mizzen, T. Stukenberg, C. D. Allis and I. G. Macara,
Quimby, B. B., C. A. Wilson and A. H. Corbett,
Sazer, S., and M. Dasso,
Sekiguchi, T., E. Hirose, N. Nakashima, M. Ii and T. Nishimoto,
Sekiguchi, T., Y. Todaka, Y. Wang, E. Hirose, N. Nakashima et al.,
Sprang, S. R.,
Tai, P. C., D. P. Kessler and J. Ingraham,
Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson et al.,
Valenzuela, P., F. Weinberg, G. Bell and W. J. Rutter,
Wang, Y., T. Sekiguchi, E. Noguchi and T. Nishimoto,
Warner, J. R.,
Yano, R., and M. Nomura,
Zanchin, N. I., and D. S. Goldfarb,
Zanchin, N. I., P. Roberts, A. DeSilva, F. Sherman and D. S. Goldfarb,



![Ribosomal RNA synthesis was decreased in the gtr1Δ strain. (a) The growth curve of NBW5 gtr1Δ (•), NBW5 gtr1Δ expressing wild-type Gtr1p (pL242) (○), NBW5 gtr1Δ expressing Q65L Gtr1p (pL244) (□), NBW5 gtr1Δ expressing S20L Gtr1p (pL243) (▵), and wild-type NBW5 (+) strains at 26° (left) and at 14° (right) was studied as previously described (Zanchin et al. 1997). Decrease in the growth rate of gtr1Δ and S20L strains was significant at 14°. (b) To examine the global rRNA synthesis rate in the strains described above, exponentially growing cultures were labeled with [3H]uracil (50 μCi/ml) for 10 min at 26° or 30 min at 14° and cold excess uracil (300 μg/ml) was added and incubated to complete the processing for 1 hr at 26° or 3 hr at 14°. Extracted RNAs were run on formaldehyde agarose gel and transferred to Hybond filters. The filters were exposed to an imaging plate (TR2040, Fuji Photo film). Lanes 1 and 5, NBW5 gtr1Δ; lanes 2 and 6, NBW5 gtr1Δ (pL242); lanes 3 and 7, NBW5 gtr1Δ (pL243); lanes 4 and 8, NBW5 gtr1Δ (pL244). Relative radioactivity (R.R.) of 25S or 18S rRNA is shown at the bottom left (gtr1Δ, 26° = 100). RNA was stained with ethidium bromide showing that an equal amount of RNA was loaded on the gel (right). (c) Cells grown at 30° to midlog phase. 5′ ITS1 rRNA was localized by fluorescence in situ hybridization. Chromosomal DNA was labeled with 4′,6-diamidino-2-phenylindole-dihydrochloride to identify the location of the nucleus. (d) Total RNA was isolated from logarithmically grown gtr1Δ, gtr2Δ, and wild-type strains at either 26° or 16° for 2 hr. Total RNA (5 μg) was run on formaldehyde agarose gels and transferred onto nylon membranes and blotted with the 32P-labeled probes indicated at the right. Relative radioactivity (R.R.) of RPS3, RPL3, or ACT1 is shown at the bottom (wild type, 26° = 100). (e) rRNA processing was examined by 2-min pulse labeling with [3H]methyl methionine and chased for 2, 6, and 15 min in the presence of methionine (100 μg/ml), as described previously (Zanchin et al. 1997). Positions of 27S, 25S, 20S, and 18S are indicated.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/genetics/170/4/10.1534_genetics.105.042366/4/m_2366f3.jpeg?Expires=1716481889&Signature=cysKuLIUMIxvYSJMdN8dH0tbJXZgGfY9pvJ7s0P6u0pgROFs2awTsJLw3wjwuTKSCMGAPsOmszdGbiKMiayF2BU-IFPTQ-b9FAqFwVXUQV4FyjU4jolYuGpxQeG7jOUXFhFaeRRSK-2W4sDcTrAkRebIpNkWyRO8xSXWK-TU7qlX27p6FWfhBVhZvTkH0iWUWZjnID5aDBv0-yj6fS4z1GILVSDtMZj1aOaTcOuR6sb-4ShbGt7GL4kFWQShnZjjob5C43F2H99Cx4PMwkGBIdFaVNNhWy7y9~PjVt7cyH1WLNpo~93zeh-73CrQl4Lz7FfEHaZklyeVJSwpaLx1eg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)