-
PDF
- Split View
-
Views
-
Cite
Cite
Carole H Sellem, Claire Lemaire, Séverine Lorin, Geneviève Dujardin, Annie Sainsard-Chanet, Interaction Between the oxa1 and rmp1 Genes Modulates Respiratory Complex Assembly and Life Span in Podospora anserina, Genetics, Volume 169, Issue 3, 1 March 2005, Pages 1379–1389, https://doi.org/10.1534/genetics.104.033837
Close - Share Icon Share
Abstract
A causal link between deficiency of the cytochrome respiratory pathway and life span was previously shown in the filamentous fungus Podospora anserina. To gain more insight into the relationship between mitochondrial function and life span, we have constructed a strain carrying a thermosensitive mutation of the gene oxa1. OXA1 is a membrane protein conserved from bacteria to human. The mitochondrial OXA1 protein is involved in the assembly/insertion of several respiratory complexes. We show here that oxa1 is an essential gene in P. anserina. The oxa1ts mutant exhibits severe defects in the respiratory complexes I and IV, which are correlated with an increased life span, a strong induction of the alternative oxidase, and a reduction in ROS production. However, there is no causal link between alternative oxidase level and life span. We also show that in the oxa1ts mutant, the extent of the defects in complexes I and IV and the life-span increase depends on the essential gene rmp1. The RMP1 protein, whose function is still unknown, can be localized in the mitochondria and/or the cytosolic compartment, depending on the developmental stage. We propose that the RMP1 protein could be involved in the process of OXA1-dependent protein insertion.
REGULATION of longevity and the processes that lead to age-related damages have been very actively studied in the past decade. Studies in model systems revealed numerous processes limiting longevity and aging. Among these processes, mitochondrial function seems to play a determinant role. The free radical theory, first proposed by Harman (1956), hypothesized that free radicals (reactive oxygen species, ROS) cause oxidative damage to proteins, lipids, and DNA, resulting in aging and death (Shigenaga et al. 1994). Mitochondria are the major source of ROS production and are also the major site of adenosine triphosphate (ATP) production. This dual role raises an endogenous challenge to the survival of the cell or organism. It is therefore not surprising that the genetic analysis of aging revealed a large number of genes essential for mitochondrial function. A causal link between respiration and life span was first established in the filamentous fungus Podospora anserina in which a loss of function of cytochrome c oxidase strikingly delays aging (Dufour et al. 2000). In the same way, several mutations that decrease the activity of the respiratory chain were shown to dramatically increase life span in Caenorhabditis elegans (Feng et al. 2001; Lee et al. 2003). Two recent studies in this organism also revealed that disturbing the synthesis of subunits of the respiratory chain by RNA interference increases life span and that genes important for mitochondrial function stand out as the principal group of genes affecting longevity (Dillin et al. 2002; Lee et al. 2003). Last but not least, a direct causal link between mitochondrial DNA mutations, respiratory chain dysfunction, and aging has just been demonstrated in mammals (Trifunovic et al. 2004). Altogether, these studies establish a strong link between low ATP content, low oxygen uptake, and increased longevity. However, the mechanisms by which mitochondrial function (ATP level, ROS level, complex metabolic changes, etc.) can regulate aging are not clearly understood.
P. anserina is a filamentous ascomycete that exhibits an unavoidable and specific arrest of vegetative growth called “senescence” (Rizet 1953). The senescent state is invariably associated with mitochondrial DNA (mtDNA) instability and accumulation of circular DNA molecules generated from the mitochondrial genome and called senDNAs (Albert and Sellem 2002 and Belcour et al. 1999 for a review). As previously shown, inactivation of cytochrome c oxidase results in an increased mtDNA stability and longevity associated with decreased ROS production and deficits in energy-consuming processes such as fertility and growth rate (Dufour et al. 2000). Absence of the cytochrome part of the respiratory chain is not lethal in P. anserina, due to the activity of an alternative oxidase (AOX) known to branch off the cytochrome pathway at ubiquinone (Vanlerberghe and McIntosh 1997; Sluse and Jarmuszkiewicz 1998 for reviews). However, the exclusive use of the alternative pathway results in the loss of two of the three potential coupling sites and a decrease in ATP synthesis efficiency. Interestingly, the constitutive expression of the AOX pathway in long-lived respiratory mutants results in an increase of both ROS and ATP production (estimated from growth improvement and fertility restoration) correlated with a reduction of longevity (Lorin et al. 2001). All of these studies clearly point to mitochondrial energy as a key determinant of life span in P. anserina but do not allow us to specify what are the mitochondrial critical parameters that control life span.
In an attempt to gain more insight into the links between mitochondrial function and life span, we have studied the role of the nuclear oxa1 gene. OXA1 is a mitochondrial membrane protein conserved from bacteria to chloroplasts and mitochondria (Kuhn et al. 2003). It is a key component of the insertion machinery of membrane proteins (Herrmann and Neupert 2003). In Saccharomyces cerevisiae, absence of Oxa1p activity is viable but mutants are unable to respire. They show severe defects in the assembly of cytochrome c oxidase (complex IV) and ATPase (complex V) associated with minor defects in the biogenesis of the bc1 complex, complex III (Bonnefoy et al. 1994a; Altamura et al. 1996; Hamel et al. 1998). In Schizosaccharomyces pombe, the absence of the OXA1 protein results in a nonviable phenotype (Bonnefoy et al. 2000). These yeasts are devoid of complex I (proton-pumping NADH dehydrogenase). Recently, it has been shown in Neurospora crassa that OXA1 is also necessary for the biogenesis of complex I and that inactivation of the gene oxa1 by the process of repeat-induced point mutation leads to lethality (Nargang et al. 2002). The demonstration that at least some complex I subunits depend on OXA1 for their assembly/insertion extends the role of OXA1 to at least four of the five respiratory complexes.
In this study, we describe the properties of a P. anserina mutant that carries a deletion of the oxa1 gene and an ectopic thermosensitive copy of the gene (Δoxa1 oxa1ts). Our results confirm that oxa1 is an essential gene in obligate aerobic fungi, even in those that possess an alternative pathway, and that the OXA1 protein plays a key role in the biogenesis of complexes I and IV. Interestingly, we found a genetic interaction between the oxa1 and rmp1 genes. The rmp1 gene exists under two natural alleles, rmp1-1 and rmp1-2, that control the timing of death in strains carrying specific mutations in the nuclear gene AS1, which encodes a cytosolic ribosomal protein (Dequard-Chablat and Sellem 1994). rmp1 is an essential gene and the RMP1 protein, whose function has not been established, can be localized in the mitochondrial and/or the cytosolic compartment, depending on the developmental stage (Contamine et al. 2004). We show here that the mutant form of OXA1 in the Δoxa1 oxa1ts strain grown at subrestrictive temperature leads to pleiotropic respiratory defects and to modification of life span. Interestingly, the extent of these alterations depends on the rmp1 gene. The significance of the interaction between the oxa1 and rmp1 genes is discussed.
MATERIALS AND METHODS
P. anserina strains, growth conditions, and transformation of protoplasts:
The genetics and biological properties of P. anserina were first described by Rizet and Engelmann (1949) and reviewed by Esser (Esser 1974). Strains used in this study were derived from the s wild-type strain (Rizet 1952). The leu1-1 mutant is auxotrophic for leucine. The gpd-aox strain contains an ectopic copy of the aox gene under the control of the strong constitutive P. anserina gpd promotor. The aox::hygro strain is inactivated for the endogenous aox gene (Lorin et al. 2001). The pHSS plasmid carries the hygromycin resistance gene and the P. anserina rmp1-1 gene (Contamine et al. 2004). All media, i.e., cornmeal extract (MR), minimal synthetic (M2), and germination media, were as described (Esser 1974). When necessary, hygromycin, phleomycin, and leucine were added to the M2 medium at a concentration of 75, 10, and 50 μg/ml, respectively. Protoplast achievement and transformation experiments were conducted as described previously (Berges and Barreau 1989), except that protoplasts were obtained by incubation in 40 mg/ml glucanex (Laffort).
Plasmids, bacterial, and yeast strains:
The plasmid used for the construction of the P. anserina cDNA libraries was the multicopy S. cerevisiae pFL61 expression vector, which carries the URA3 gene as selectable marker in yeast (d'Enfert et al. 1995). The plasmids used for constructions of the P. anserina strains bearing a deleted oxa1 gene (Δoxa1) and/or an ectopic thermosensitive oxa1 allele (oxa1ts) were pBC-hygro, which carries the bacterial hph gene conferring resistance to hygromycin as selectable marker (Silar 1995), and pUL, which carries the wild-type P. anserina leu1 gene (Turcq 1989; Debuchy et al. 1993). The TOPO vector (Invitrogen, San Diego) was used to clone PCR amplification products. The pPable plasmid is a pUC vector containing the Tn5 bacterial bleomycin (ble) gene under the control of the P. anserina gpd promotor and the S. cerevisiae cyc1 terminator (Coppin and Debuchy 2000). The ble gene confers resistance to phleomycin. Plasmids were propagated in Escherichia coli XL1blue. Growth culture conditions and genetic and transformation methods for S. cerevisiae were as reported (Bonnefoy et al. 1994a).
Construction of P. anserina cDNA libraries in the expression vector of yeast pFL61:
Total RNA (2 mg) was extracted from the s wild-type strain (mat−) and ∼20 μg of poly(A+) RNA was purified twice on oligo(dT)-cellulose columns (mRNA purification kit, Pharmacia). Anchored dT25 primers were used to obtain 3.2 μg double-strand DNA (cDNA kit, Amersham, Buckinghamshire, UK). Three cDNA libraries, corresponding to three ranges of molecular weight cDNA (0.2–1 kb, 1–2.5 kb, and >2.5 kb) were cloned using BstX1 adaptators in the pFL61 vector between the 5′ (promoter) and 3′ (terminator) sequences of the pgk1 gene as described previously (d'Enfert et al. 1995). Sequencing of these cDNA libraries is currently in progress at Genoscope (Evry, France).
Complementation of the yeast NBT2 mutant strain and isolation of the oxa1 cDNA:
Since the pFL61 plasmid carries the URA3 gene, the transformation events of the NBT2 strain—oxa1::LEU2, leu2-3,112, ura3-1, his3-11,15, trp1-1, ade2-1, mat α (Hamel et al. 1998)—can be easily selected by plating on a fermentable medium (glucose) lacking uracil and containing histidine, tryptophan, and adenine. Among >105 transformants screened, 50 and 5 transformants able to grow on nonfermentable carbon source (glycerol) were obtained from the 0.2- to 1-kb and the 1- to 2.5-kb cDNA libraries, respectively. Five of them were analyzed. They all contained a 1.2-kb insert whose sequence was identical and whose translated product is homologous to the OXA1 protein of several organisms.
Cloning, sequencing, and localization of the P. anserina oxa1 gene:
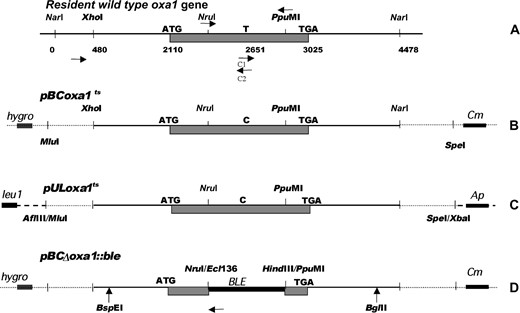
The oxa1 cDNA used as a probe in Southern blot analysis revealed a 4.5-kb NarI DNA genomic fragment. A NarI mini-bank was thus constructed and screened with the same probe. The sequence of the 4478-bp cloned NarI fragment (Figure 1A)
Cloning and inactivation of the oxa1 gene. (A) Partial restriction map of the P. anserina wild-type oxa1 gene. Nomenclature is according to the submitted sequence (no. AF 382189). The arrows represent the oligonucleotides used for further PCR experiments with their 5′–3′orientation. (B) Partial restriction map of the pBCoxa1ts vector. The XhoI-NarI fragment of the P. anserina wild-type oxa1 gene was cloned in the pBC-hygro vector (pBCoxa1). An NruI-PpuMI modified fragment obtained in a two-step PCR amplification replaced the NruI-PpuMI wild-type fragment. The first step was the amplification of two fragments using two couples of primers (NruI/C2 and C1/PpuMI), including two complementary oligonucleotides (C1 and C2) designed with the desired T-to-C substitution. In the second step, the NruI/PpuMI fragment was amplified using the two preceding amplified fragments as matrixes. (C) Partial restriction map of the pULoxa1ts vector. The MluI-SpeI fragment of pBCoxa1ts was cloned in a pUL vector carrying the leu1 gene. (D) Partial restriction map of the pBCΔoxa1::ble vector. The NruI-PpuMI wild-type fragment of pBCoxa1 was replaced by an Ecl136II-HindIII DNA fragment carrying the ble gene using a HindIII/PpuMI linker. The shaded boxes correspond to the oxa1 coding sequence. Dashed and dotted lines correspond to plasmid sequences. Ap, Cm, and hygro are bacterial genes conferring, respectively, the ampicillin, chloramphenicol, and hygromycin resistance to the pUC- and pBC-hygro-derived vectors.
was determined and registered with the accession no. AF382189. A shorter (4-kb) XhoI-NarI fragment was subcloned in the pBC-hygro vector digested by XhoI and ClaI. The resulting pBCoxa1 plasmid was further used for various subclonings. The oxa1 gene was located by hybridization on the separated chromosomes of P. anserina slotted on a nylon membrane (Barreau et al. 2002) kindly provided by C. Barreau using the oxa1 cDNA as a probe (chromoslot technique).
Construction of strains containing an ectopic copy of the oxa1 gene carrying a L269S substitution:
The construction of a transformation vector containing a thermosensitive oxa1 allele was achieved as shown in Figure 1. First, a T-to-C transition in position 2651 was introduced in the open reading frame of the wild-type oxa1 gene of pBCoxa1 to obtain a L269S substitution located in the loop between the transmembrane domains 2 and 3 of the OXA1 protein. This change corresponded to the L240S substitution introduced into the S. cerevisiae oxa1 gene and resulted in the thermosensitive pet1402 mutant (Bauer et al. 1994; Meyer et al. 1997a). This modification was achieved by direct mutagenesis in a two-step PCR amplification of the internal NruI-PpuMI DNA fragment (see legend of Figure 1B) and it was ascertained by sequencing. Second, the NruI-PpuMI of pBCoxa1 was excised and replaced by the modified NruI-PpuMI DNA to give pBCoxa1ts (Figure 1B). Third, a 5.7-kb DNA fragment (MluI-SpeI) from pBCoxa1ts containing the L269S allele was subcloned in the pUL vector digested by AflIII and XbaI. The resulting pULoxa1ts vector (Figure 1C) was further used to transform the leu1-1 strain to obtain ectopic integrations of the oxa1ts gene (L269S oxa1 allele). Five independent ectopic integration events of the plasmid were selected on minimal medium. These five primary transformants were crossed with the leu1-1 strain to obtain prototroph monocaryotic transformed strains carrying the resident oxa1+ gene and an ectopic L269S oxa1 copy associated with both mating types. The oxa1+ (oxa1ts) resulting strain (the transgene in parentheses) had no observable phenotype and showed the same growth rate, mycelium aspect, and fertility as the wild-type strain at 27° as at 34°.
Inactivation of the oxa1 gene and construction of strains bearing the Δoxa1 allele:
The construction of the pBCΔoxa1::ble plasmid used to obtain a null allele of gene oxa1 was achieved as follows: the internal NruI-PpuMI fragment of pBCoxa1 that contained most of the coding sequence of gene oxa1 was excised and replaced by a 1.1-kb DNA fragment carrying the phleomycin resistance gene (ble). This construction gave the pBCΔoxa1::ble vector (Figure 1D). A linear DNA fragment (BspEI-BglII) of 3.7 kb issued from the pBCΔoxa1::ble vector containing 1.4 kb upstream from the NruI site and 1.2 kb downstream from the PpuMI site was used to transform a leu1-1 mat+ strain carrying an oxa1ts ectopic copy. A total of 200 transformants, screened for phleomycin resistance, were tested by PCR analysis for the replacement of the wild-type oxa1 resident gene by the inactivated Δoxa1 allele. For this purpose, a couple of primers designed in the genomic sequence, upstream from the XhoI restriction site (sense) and in the ble gene (antisense) were used (Figure 1, A and D). PCR amplifications were performed directly on crude extracts of lyophilized mycelia. Twelve transformants revealed a modified phenotype at 28° and 32° and 2 were slow growing only at 32°. One of the latter allowed the amplification of a fragment of the expected size.
Isolation of mitochondria:
Mitochondria from cultures grown at 27° or 30° were isolated by differential centrifugation as described previously (Sellem et al. 1990). Proteins were resuspended in 0.7 m sorbitol, 50 mm Tris-HCl, pH 7.5, and 0.2 mm EDTA containing protease inhibitor and further used for Western blot or BN-PAGE analysis. Bio-Rad (Richmond, CA) assay was used to determine protein concentration.
RESULTS
Characteristics of the oxa1 sequence:
Cloning of the P. anserina oxa1 cDNA by functional complementation in a S. cerevisiae oxa1 null mutant revealed that the P. anserina OXA1 protein can rescue the yeast respiratory deficiency. The 426-amino-acid protein deduced from the P. anserina oxa1 sequence shares 41, 21, and 22% identity with the OXA1 proteins from N. crassa, S. cerevisiae, and Homo sapiens, respectively. The same hydrophobic distribution of amino acids revealed the same characteristic structure with a conserved core region that probably also contains five transmembrane domains. A strong coiled-coil (α-helix) structure, proposed to play a role in the recruitment of the mitochondrial translation machinery (Szyrach et al. 2003), was predicted at the C-terminal domain of the P. anserina protein as in that of N. crassa and S. cerevisiae. The oxa1 gene was cloned and modified as described in materials and methods (see Figure 1). The phleomycin-resistant phenotype associated with the oxa1 inactivation allowed us to genetically localize the oxa1 gene far from its centromere (second-division segregation percentage of ∼90). It was assigned to chromosome II by the chromoslot technique.
The inactivation of oxa1 is lethal in P. anserina:
A mat+ strain bearing the Δoxa1 allele was obtained as described in materials and methods. The recipient strain used for the transformation-mediated gene replacement carried an ectopic copy of the oxa1 gene with an L269S substitution (oxa1ts). The primary transformant strain, Δoxa1 mat+(oxa1ts), was crossed at 27° with a leu1-1 mat− strain. In the progeny, we looked for asci-containing ascospores bearing the Δoxa1 allele (determined by resistance to phleomycin) without the oxa1ts ectopic copy (determined by auxotrophy for leucine). Among ∼50 asci analyzed (each containing two monocaryotic spores), 8 asci revealed nonambiguously that homocaryotic spore resistant for phleomycin and auxotroph for leucine were unable to germinate. The fact that all germinating spores of the phleomycin-resistant phenotype are also leucine prototrophs indicates that they can survive only when the oxa1ts transgene covers for the deleted allele and allows us to conclude that the inactivation of the oxa1 gene is lethal for P. anserina.
An L269S substitution in the gene oxa1 confers a thermosensitive phenotype with pleiotropic effects and modified longevity at subrestrictive temperature:
The Δoxa1 (oxa1ts) mutant was grown at several temperatures ranging from 27° to 34°, and the life span, growth rate, and female fertility were analyzed. As shown in Table 1
Growth characteristics of the strains at permissive (27°), subrestrictive (30°), and restrictive (34°) temperatures
. | Lifespan (cm) . | Growth rate (cm/day) . | Female fertility . | |||||
|---|---|---|---|---|---|---|---|---|
| Strain . | 27° . | 30° . | 34° . | 27° . | 30° . | 34° . | 27° . | 30° . |
| Wild-type mat− | 8.1 ± 1.3 (6) | 9.1 ± 1.7 (9) | 7.0 ± 1.5 (3) | 0.82 ± 0.05 | 0.81 ± 0.09 | 0.90 ± 0.11 | 100 | 100 |
| Wild-type mat+ | 9.1 ± 3.7 (7) | 10.8 ± 2.5 (6) | 5.1 ± 2.6 (3) | 0.80 ± 0.06 | 0.80 ± 0.03 | 0.89 ± 0.22 | 100 | 100 |
| Δoxa1 (oxa1ts) mat− | 5.8 ± 2.2 (13) | 5.1 ± 1.8 (15) | − | 0.80 ± 0.10 | 0.75 ± 0.05 | − | 38 | 8 |
| Δoxa1 (oxa1ts) mat+ | 9.3 ± 3.8 (13) | 28.1 ± 17.0 (17) | − | 0.77 ± 0.08 | 0.70 ± 0.09 | − | 27 | 5 |
| Δoxa1 (oxa1ts) (rmp1-1) mat− | 6.1 ± 0.5 (2) | 5.2 ± 0.5 (2) | ||||||
| Δoxa1 (oxa1ts) (rmp1-1) mat+ | 4.2 ± 2.5 (4) | 4.6 ± 0.5 (5) | ||||||
| Δoxa1 (oxa1ts) (gpd-aox) mat− | 10.5 ± 3.4 (13) | 10.6 ± 3.3 (5) | ||||||
| Δoxa1 (oxa1ts) (gpd-aox) mat+ | 11.6 ± 3.5 (14) | 43.2 ± 23.6 (10) | ||||||
| Δoxa1 (oxa1ts) (Δaox) mat− | 7.0 ± 0.5 (3) | 7.0 ± 0.7 (3) | ||||||
| Δoxa1 (oxa1ts)
(Δaox) mat+ | 8.7 ± 1.2 (3) | 28.1 ± 20.7 (14) | ||||||
. | Lifespan (cm) . | Growth rate (cm/day) . | Female fertility . | |||||
|---|---|---|---|---|---|---|---|---|
| Strain . | 27° . | 30° . | 34° . | 27° . | 30° . | 34° . | 27° . | 30° . |
| Wild-type mat− | 8.1 ± 1.3 (6) | 9.1 ± 1.7 (9) | 7.0 ± 1.5 (3) | 0.82 ± 0.05 | 0.81 ± 0.09 | 0.90 ± 0.11 | 100 | 100 |
| Wild-type mat+ | 9.1 ± 3.7 (7) | 10.8 ± 2.5 (6) | 5.1 ± 2.6 (3) | 0.80 ± 0.06 | 0.80 ± 0.03 | 0.89 ± 0.22 | 100 | 100 |
| Δoxa1 (oxa1ts) mat− | 5.8 ± 2.2 (13) | 5.1 ± 1.8 (15) | − | 0.80 ± 0.10 | 0.75 ± 0.05 | − | 38 | 8 |
| Δoxa1 (oxa1ts) mat+ | 9.3 ± 3.8 (13) | 28.1 ± 17.0 (17) | − | 0.77 ± 0.08 | 0.70 ± 0.09 | − | 27 | 5 |
| Δoxa1 (oxa1ts) (rmp1-1) mat− | 6.1 ± 0.5 (2) | 5.2 ± 0.5 (2) | ||||||
| Δoxa1 (oxa1ts) (rmp1-1) mat+ | 4.2 ± 2.5 (4) | 4.6 ± 0.5 (5) | ||||||
| Δoxa1 (oxa1ts) (gpd-aox) mat− | 10.5 ± 3.4 (13) | 10.6 ± 3.3 (5) | ||||||
| Δoxa1 (oxa1ts) (gpd-aox) mat+ | 11.6 ± 3.5 (14) | 43.2 ± 23.6 (10) | ||||||
| Δoxa1 (oxa1ts) (Δaox) mat− | 7.0 ± 0.5 (3) | 7.0 ± 0.7 (3) | ||||||
| Δoxa1 (oxa1ts)
(Δaox) mat+ | 8.7 ± 1.2 (3) | 28.1 ± 20.7 (14) | ||||||
To determine life span, cultures were grown on race tubes containing minimal synthetic M2 medium with twice the usual amount of agar at various temperatures ranging from 27° to 34°. Life spans (± standard deviation) are mean values determined in centimeters of growth initiated from the ascospore to the arrested edge of 2–17 parallel cultures of 1–10 different ascospores of the same genotype (the total number of cultures is given in parentheses). −, absence of growth. For each strain, growth rate was estimated on 5–15 subcultures over 5 days of growth and expressed in centimeters of growth per day. Female fertility was estimated by the number of mature perithecia that developed when the strains were fertilized with a suspension of wild-type conidia of the opposite mating type and is expressed as the percentage of the fertility of the wild-type strain of the same mating type at the same temperature. Transgenes are indicated in parentheses. The resident rmp1 allele is not indicated in the table: all the mat− strains are rmp1-1; all the mat+ strains are rmp1-2.
Growth characteristics of the strains at permissive (27°), subrestrictive (30°), and restrictive (34°) temperatures
. | Lifespan (cm) . | Growth rate (cm/day) . | Female fertility . | |||||
|---|---|---|---|---|---|---|---|---|
| Strain . | 27° . | 30° . | 34° . | 27° . | 30° . | 34° . | 27° . | 30° . |
| Wild-type mat− | 8.1 ± 1.3 (6) | 9.1 ± 1.7 (9) | 7.0 ± 1.5 (3) | 0.82 ± 0.05 | 0.81 ± 0.09 | 0.90 ± 0.11 | 100 | 100 |
| Wild-type mat+ | 9.1 ± 3.7 (7) | 10.8 ± 2.5 (6) | 5.1 ± 2.6 (3) | 0.80 ± 0.06 | 0.80 ± 0.03 | 0.89 ± 0.22 | 100 | 100 |
| Δoxa1 (oxa1ts) mat− | 5.8 ± 2.2 (13) | 5.1 ± 1.8 (15) | − | 0.80 ± 0.10 | 0.75 ± 0.05 | − | 38 | 8 |
| Δoxa1 (oxa1ts) mat+ | 9.3 ± 3.8 (13) | 28.1 ± 17.0 (17) | − | 0.77 ± 0.08 | 0.70 ± 0.09 | − | 27 | 5 |
| Δoxa1 (oxa1ts) (rmp1-1) mat− | 6.1 ± 0.5 (2) | 5.2 ± 0.5 (2) | ||||||
| Δoxa1 (oxa1ts) (rmp1-1) mat+ | 4.2 ± 2.5 (4) | 4.6 ± 0.5 (5) | ||||||
| Δoxa1 (oxa1ts) (gpd-aox) mat− | 10.5 ± 3.4 (13) | 10.6 ± 3.3 (5) | ||||||
| Δoxa1 (oxa1ts) (gpd-aox) mat+ | 11.6 ± 3.5 (14) | 43.2 ± 23.6 (10) | ||||||
| Δoxa1 (oxa1ts) (Δaox) mat− | 7.0 ± 0.5 (3) | 7.0 ± 0.7 (3) | ||||||
| Δoxa1 (oxa1ts)
(Δaox) mat+ | 8.7 ± 1.2 (3) | 28.1 ± 20.7 (14) | ||||||
. | Lifespan (cm) . | Growth rate (cm/day) . | Female fertility . | |||||
|---|---|---|---|---|---|---|---|---|
| Strain . | 27° . | 30° . | 34° . | 27° . | 30° . | 34° . | 27° . | 30° . |
| Wild-type mat− | 8.1 ± 1.3 (6) | 9.1 ± 1.7 (9) | 7.0 ± 1.5 (3) | 0.82 ± 0.05 | 0.81 ± 0.09 | 0.90 ± 0.11 | 100 | 100 |
| Wild-type mat+ | 9.1 ± 3.7 (7) | 10.8 ± 2.5 (6) | 5.1 ± 2.6 (3) | 0.80 ± 0.06 | 0.80 ± 0.03 | 0.89 ± 0.22 | 100 | 100 |
| Δoxa1 (oxa1ts) mat− | 5.8 ± 2.2 (13) | 5.1 ± 1.8 (15) | − | 0.80 ± 0.10 | 0.75 ± 0.05 | − | 38 | 8 |
| Δoxa1 (oxa1ts) mat+ | 9.3 ± 3.8 (13) | 28.1 ± 17.0 (17) | − | 0.77 ± 0.08 | 0.70 ± 0.09 | − | 27 | 5 |
| Δoxa1 (oxa1ts) (rmp1-1) mat− | 6.1 ± 0.5 (2) | 5.2 ± 0.5 (2) | ||||||
| Δoxa1 (oxa1ts) (rmp1-1) mat+ | 4.2 ± 2.5 (4) | 4.6 ± 0.5 (5) | ||||||
| Δoxa1 (oxa1ts) (gpd-aox) mat− | 10.5 ± 3.4 (13) | 10.6 ± 3.3 (5) | ||||||
| Δoxa1 (oxa1ts) (gpd-aox) mat+ | 11.6 ± 3.5 (14) | 43.2 ± 23.6 (10) | ||||||
| Δoxa1 (oxa1ts) (Δaox) mat− | 7.0 ± 0.5 (3) | 7.0 ± 0.7 (3) | ||||||
| Δoxa1 (oxa1ts)
(Δaox) mat+ | 8.7 ± 1.2 (3) | 28.1 ± 20.7 (14) | ||||||
To determine life span, cultures were grown on race tubes containing minimal synthetic M2 medium with twice the usual amount of agar at various temperatures ranging from 27° to 34°. Life spans (± standard deviation) are mean values determined in centimeters of growth initiated from the ascospore to the arrested edge of 2–17 parallel cultures of 1–10 different ascospores of the same genotype (the total number of cultures is given in parentheses). −, absence of growth. For each strain, growth rate was estimated on 5–15 subcultures over 5 days of growth and expressed in centimeters of growth per day. Female fertility was estimated by the number of mature perithecia that developed when the strains were fertilized with a suspension of wild-type conidia of the opposite mating type and is expressed as the percentage of the fertility of the wild-type strain of the same mating type at the same temperature. Transgenes are indicated in parentheses. The resident rmp1 allele is not indicated in the table: all the mat− strains are rmp1-1; all the mat+ strains are rmp1-2.
, this mutant cannot grow at 34°, and 31° was the highest temperature compatible with growth indicating that, as in S. cerevisiae, the L269S substitution confers a thermosensitive phenotype. Life-span determination revealed no effect of the mutation at 27° whereas at the subrestrictive temperature of 30°, the Δoxa1 (oxa1ts) mat− cultures displayed a reduced life span and the Δoxa1 (oxa1ts) mat+ cultures an increased one (at least 250% of the wild type). The life span of Δoxa1 (oxa1ts) mat+ cultures appeared extremely heterogenous (see the standard deviation in Table 1) and the arrests of growth did not correspond to death of the mycelium, which always resumed growth after transfer to 27°. In contrast, the arrests of growth at 27° and 30° of the Δoxa1 (oxa1ts) mat− and at 27° of the Δoxa1 (oxa1ts) mat+ strains were due to death and the apical cells were unable to grow when transferred at 27°. The growth rate was almost unaffected at 27° in Δoxa1 (oxa1ts) strains, irrespective of the mating type, and only slightly reduced for the Δoxa1 (oxa1ts) mat+ strain grown at 30°. In contrast, female fertility appeared very reduced even at 27° and this defect was much more marked at 30° where female fertility of the Δoxa1 (oxa1ts) strains was <10% of that of the wild-type strain.
Growth arrest in the wild-type strain is due to senescence and systematically correlated with the accumulation of circular, tandemly arranged mtDNA called senDNAs (Belcour et al. 1999). Whereas senDNAs were revealed in Δoxa1 (oxa1ts) mat− and mat+ cultures dying at 27° and in Δoxa1 (oxa1ts) mat− cultures dying at 30°, arrest of growth of the Δoxa1 (oxa1ts) mat+ cultures at 30° was not accompanied with such accumulation of senDNAs (data not shown). This result favors the idea that the arrest of growth in the cultures with increased life span (Δoxa1 (oxa1ts) mat+ at 30°) is not the result of a senescence process.
Gene oxa1 interacts with the nuclear mitochondrial gene rmp1:
In P. anserina, a gene closely linked to the mat locus (<0.5 cM) has been identified (Contamine et al. 1996, 2004). This gene, rmp1, exists under two natural alleles in strain s: rmp1-1 and rmp1-2, linked, respectively, to the mat− and mat+ loci. Gene rmp1 controls the timing of premature death, a syndrome characterized by the accumulation of specific mitochondrial deletions (Belcour et al. 1991). This syndrome is observed only in strains carrying some mutations in the AS1 gene, such as AS1-4, which alter a cytosolic ribosomal protein (Dequard-Chablat et al. 1986). Both AS1-4 rmp1-1 and AS1-4 rmp1-2 strains accumulate specific mitochondrial deletions at time of death, but they strongly differ in their longevity, which is ∼2 and 80 cm, respectively (Contamine et al. 1996). Gene rmp1 has been shown to be an essential gene encoding a protein that can be localized in the mitochondrial and/or the cytosolic compartments (Contamine et al. 2004). So, we asked whether the differences observed between Δoxa1 (oxa1ts) mat+ and mat− strains at 30° revealed an interaction between the oxa1 gene and the mat locus or between the oxa1 gene and the rmp1 gene. To address this question, we took advantage of the cloning of the rmp1-1 allele on an hygromycin transformation vector and of the dominance of this allele (Contamine et al. 2004) to introduce rmp1-1 by transformation into a Δoxa1 (oxa1ts) rmp1-2 mat+ strain. Secondary Δoxa1 (oxa1ts) (rmp1-1) rmp1-2 mat+ transformants were obtained and analyzed. Interestingly, as shown in Table 1, this analysis revealed that, when introduced into a Δoxa1 (oxa1ts) rmp1-2 mat+ strain, rmp1-1 leads to a reduced life span, not different from that of Δoxa1 (oxa1ts) rmp1-1 mat−, and to a restoration of accumulation of senDNA at the time of death (not shown). These data clearly demonstrate that the timing of death and the instability of the mtDNA in the Δoxa1 (oxa1ts) strains grown at subrestrictive temperature depend on the rmp1 gene and not on the mat locus.
The activity of the respiratory complexes I and IV is differentially impaired in Δoxa1 (oxa1ts) rmp1-1 and rmp1-2 strains:
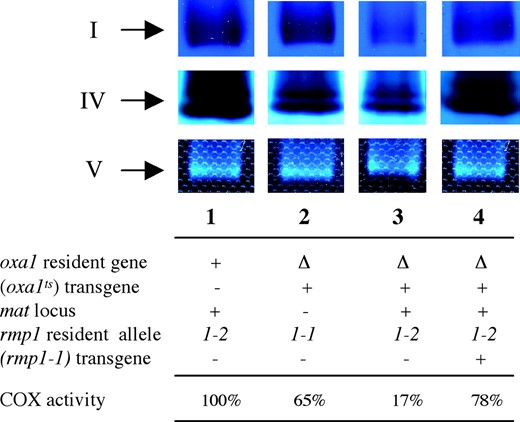
As demonstrated in several organisms, the OXA1 protein is required for the biogenesis of several respiratory complexes, namely complexes IV and V (Bonnefoy et al. 1994a; Altamura et al. 1996) and complex I (Nargang et al. 2002). The effect of the oxa1ts mutation on the activity of these different complexes was investigated in P. anserina Δoxa1 (oxa1ts) strains grown at 30°. Blue-native polyacrylamide gel electrophoresis (BN-PAGE), which allows a one-step separation of the different complexes in a gel activity assay, was performed (Figure 2)
In-gel detection of complex I, IV, and V activities in various strains grown at 30°. Sample preparation and blue-native polyacrylamide gel electrophoresis (BN-PAGE) were carried out as described (Schagger and von Jagow 1991). Mitochondria from P. anserina were rendered soluble in the presence of 2% (w/v) N-dodecyl maltoside. In-gel activity assays of complexes I, IV, and V were performed as described (Nijtmans et al. 2002). Genotypes of the strains are detailed below the gel. The oxa1 gene is either present (+) or inactivated (Δ). + or − indicates the presence or the absence of the (oxa1ts) and (rmp1-1) transgenes. The two mat alleles, mat+ and mat−, are indicated by + and −, respectively. The specific activity of cytochrome c oxidase (COX) is expressed as the percentage of the wild type (wild type is 856 ± 54 nmol of oxidized cytochrome c per minute per milligram of mitochondrial protein). The wild-type strain (lane 1) gave the same results irrespective of mat and rmp1 haplotype.
. As expected from the corresponding S. cerevisiae pet1402 mutant (Meyer et al. 1997a), complex V did not seemed to be significantly affected in P. anserina Δoxa1 (oxa1ts) strains despite the information at or near the mat haplotype (lanes 2 and 3). In the same extracts a very clear difference was observed between Δoxa1 (oxa1ts) rmp1-1 mat− and Δoxa1 (oxa1ts) rmp1-2 mat+ for complex I assembly/activity, which was only slightly affected, if at all, in the rmp1-1 mat− strain (lane 2) or, in contrast, strongly reduced in the rmp1-2 mat+ strain (lane 3). In the same way, it seemed that complex IV assembly/activity was lower in Δoxa1 (oxa1ts) rmp1-2 mat+ than in Δoxa1 (oxa1ts) rmp1-1 mat− strains. To verify this difference, specific activities of complex IV were determined on isolated mitochondria. They were evaluated to correspond, respectively, to ∼20% for rmp1-2 mat+ and 70% for rmp1-1 mat− of the wild-type strain. Interestingly, the analysis of the Δoxa1 (oxa1ts) rmp1-2 mat+ strain containing the rmp1-1 transgene (lane 4) revealed a restoration of both complexes I and IV, indicating that in P. anserina the biogenesis of these complexes depends on both OXA1 and RMP1 proteins.
The alternative pathway is differentially induced in Δoxa1 (oxa1ts) rmp1-1 and rmp1-2 strains:
It is well established that absence of the Oxa1p function leads to a drastic reduction in complex IV activity in S. cerevisiae and S. pombe and that decreased OXA1 level leads to reduced content in some subunits of this complex in N. crassa (Nargang et al. 2002). The results presented here demonstrate that in P. anserina a decreased activity of complex IV is also observed in Δoxa1 (oxa1ts) strains. In this organism, impairment in complex IV is known to cause the induction of the alternative pathway, whose expression is practically null in wild type (Dufour et al. 2000; Borghouts et al. 2001; Lorin et al. 2001).
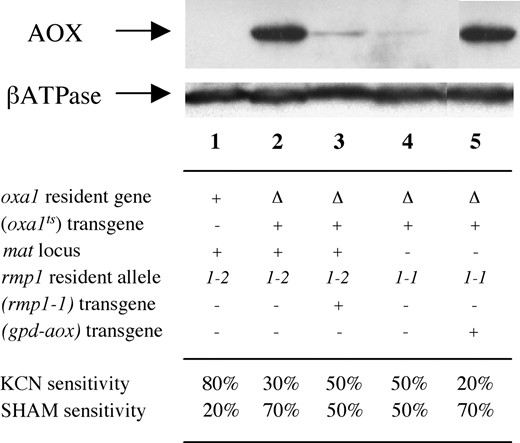
To investigate whether the alternative oxidase is induced in the Δoxa1 (oxa1ts) strains, immunodetection of the AOX protein by Western blot analyses was performed. The analysis revealed that when mitochondria were extracted from cultures grown at 27°, no AOX could be detected in the wild-type or the mutant Δoxa1 (oxa1ts) mat+ rmp1-2 and mat− rmp1-1 strains. In contrast, as shown in Figure 3
Immunochemical detection of AOX by Western blot analysis. Twenty micrograms of mitochondrial proteins extracted from cultures grown at 30° were fractionated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were probed first with an anti-AOX mouse monoclonal antibody generated against the AOX of Sauromatum guttatum (Elthon et al. 1989). Blots were then reprobed with an anti-βATPase rabbit antibody as a standardization control. The bound antibodies were detected using an enhanced chemiluminescence detection system (Pierce, Rockford, IL). For genotypes see legend of Figure 2. The percentage of respiration sensitive to cyanide (KCN, 1 mm) and SHAM (1 mg/ml) reflects the engagement of electrons in the cytochrome and alternative pathway, respectively. Oxygen uptake measurements were performed on protoplast suspensions (108 protoplasts/ml in 0.6 m sorbitol) in an oxytherm chamber with a Clark-type O2 electrode (Hansatech). Wild-type strains used as controls (lane 1) gave the same results irrespective of their mat and rmp1 haplotype.
, when mitochondria were extracted from cultures grown at 30°, there was no detectable protein in the wild type (lane 1) whereas a very low or a high amount of the AOX protein was present in Δoxa1 (oxa1ts) rmp1-1 mat− (lane 4) and rmp1-2 mat+ (lane 2), respectively. The level of induction of AOX in the Δoxa1 (oxa1ts) rmp1-2 strains at 30° was estimated to be equivalent to that observed in strains containing the aox gene under the transcriptional control of the strong constitutive gpd promoter (lane 5). It is worth noting that, as for the other parameters tested, introduction of rmp1-1 in Δoxa1 (oxa1ts) rmp1-2 mat+ restored a Δoxa1 (oxa1ts) rmp1-1 mat− phenotype and led to a spectacular decrease in the AOX level (lane 3).
To ensure that the alternative pathway induced in Δoxa1 (oxa1ts) strains is functional, the relative extent to which specific inhibitors of the alternative (salicylhydroxamic acid, SHAM) or cytochromic (potassium cyanide, KCN) pathways inhibit oxygen uptake was measured. As shown at the bottom of Figure 3, respiration of the wild type grown at 30° was ∼80% and 20% sensitive to KCN and SHAM, respectively. In contrast, respiration of the Δoxa1 (oxa1ts) strains was more sensitive to SHAM and less sensitive to KCN and this particularity was much more marked for the Δoxa1 (oxa1ts) rmp1-2 mat+ strains whose respiration was ∼30% and 70% sensitive to KCN and SHAM, respectively. This clearly indicated that, in the Δoxa1 (oxa1ts) rmp1-2 mat+ strains grown at subrestrictive temperature, the alternative pathway was the most important way of transferring the electrons to oxygen. These results are in agreement with the Western blot analysis and it can be concluded that there is either a very strong or a faint induction of the alternative pathway in the Δoxa1 (oxa1ts) strains grown at subrestrictive temperature according to the rmp1 haplotype.
The level of expression of the alternative oxidase does not causally control the phenotypic differences between Δoxa1 (oxa1ts) rmp1-1 and rmp1-2:
Relationship between life span and the alternative oxidase level in cytochrome-deficient P. anserina mutants is far from clear. So we decided to test whether or not the differences in life span observed between the Δoxa1 (oxa1ts) rmp1-1 and rmp1-2 strains are causally related to the level of expression of the AOX protein. First, to address this question, Δoxa1 (oxa1ts) strains carrying the aox inactivated gene (aox::hygro) or an ectopic copy of the aox gene under the control of the strong constitutive gpd promoter (gpd-aox) were constructed by crossing Δoxa1 (oxa1ts) with the aox::hygro (Δaox) and the gpd-aox strain (Lorin et al. 2001), respectively. The analysis of these crosses first revealed that Δoxa1 (gpd-aox) spores (determined by phleomycin and hygromycin resistance) did not germinate and that Δoxa1 (oxa1ts) (gpd-aox) rmp1-2 mat+ and rmp1-1 mat− strains did not grow beyond 31°, indicating that the constitutive expression of the gpd-aox gene does not rescue the lethal phenotype conferred by the oxa1 gene inactivation. Second, the determination of the life span of the strains carrying the gpd-aox transgene revealed, as shown in Table 1, that although the constitutive expression of AOX seemed to slightly improve life span in both Δoxa1 (oxa1ts) (gpd-aox) rmp1-1 mat− and rmp1-2 mat+ strains, it did not confer to Δoxa1 (oxa1ts) (gpd-aox) rmp1-1 mat− strains the long life span of Δoxa1 (oxa1ts) rmp1-2 mat+ strains. We have verified that the gpd-aox transgene was efficiently expressed in the Δoxa1 (oxa1ts) (gpd-aox) rmp1-1 mat− strain, as shown in Figure 3 (lane 5). The functionality of the gpd-aox transgene is attested to by the increased sensitivity to SHAM (70%). Third, as shown in Table 1, the inactivation of the aox gene in Δoxa1 (oxa1ts) rmp1-2 mat+ Δaox strains did not significantly reduce life span. Altogether, these data indicate that the different level of expression of the AOX protein between Δoxa1 (oxa1ts) rmp1-2 mat+ and rmp1-1 mat− strains at 30° is not the causal factor that accounts for the difference in longevity between the two strains.
ROS production is lowered in Δoxa1 (oxa1ts) rmp1-2 strains:
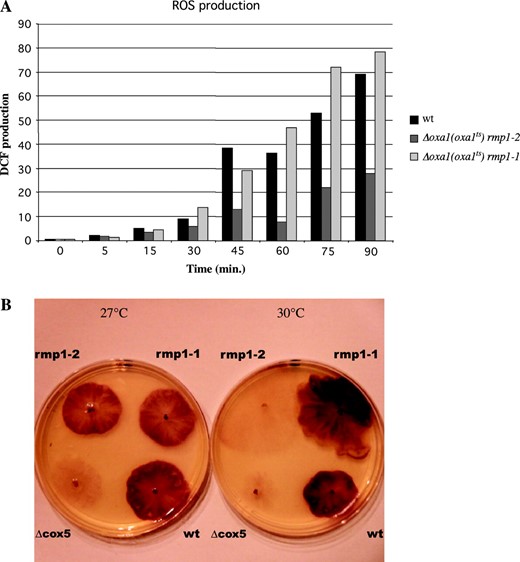
In P. anserina, absence of complex IV activity results in an increased life span and a decreased production of reactive oxygen species (Dufour et al. 2000). To test whether there is a correlation between life span and production of ROS in the Δoxa1 (oxa1ts) strains, ROS production was measured by cytofluorometry on protoplasts harvested from cultures grown at 30°. Figure 4A
ROS measurements. (A) ROS production estimated as the kinetics of dichorofluorescin (DCF) production resulting from the oxidation of the diacetate form (H2DCF-DA) as described previously (Dufour et al. 2000). For each strain grown at 30°, ∼107 protoplasts were incubated with H2DCF-DA (80 μm in 0.6 m sortitol/10 mm Tris-HCl pH 7.5) at 30° and ROS production was analyzed by measuring, every 10 min, the apparition of the fluorescent DCF in a XL3C flow cytofluometer (Coulter, France). (B) Histochemical detection of superoxide radicals on mycelium (Munkres 1990). Cultures were grown at 27° or 30° on M2 medium until a diameter of 3 cm was reached: the petri dishes were flooded with 3–5 ml of staining solution containing 2.5 mm NBT diluted in 5 mm 3-(N-morpholino) propane sulfonate-NaOH (MOPS) buffer, pH 7.6. After an incubation of 60 min at 30°, the solution was discarded and plates kept in the dark. A blue, water-insoluble formazan precipitate, obtained from the reduction of the water-soluble NBT, visualized superoxide radicals. rmp1-1, rmp1-2, Δcox5, and wt, respectively, correspond to Δoxa1 (oxa1ts) rmp1-1 mat−, Δoxa1 (oxa1ts) rmp1-2 mat+, Δcox5 mat+, and the wild-type mat+ strains. These last two strains gave the same results irrespective of their mat and rmp1 haplotype.
shows the results of three independent experiments indicating that the Δoxa1 (oxa1ts) rmp1-2 mat+ strain produced 50–60% less ROS than the wild type did while the Δoxa1 (oxa1ts) rmp1-1 mat− strain produced as much or more. These data are in good agreement with the histochemical superoxide radical detection shown in Figure 4B. In this test, the relative level of produced superoxide radicals was visualized by the relative ability of mycelia to reduce yellow water-soluble nitro-blue tetrazolium (NBT) to blue water-insoluble formazan (Munkres 1990). The amount of secreted superoxide radicals seems to be equivalent in wild-type and Δoxa1 (oxa1ts) rmp1-2 mat+ and rmp1-1 mat− strains grown at 27°. In contrast, it is dramatically reduced in the Δoxa1 (oxa1ts) rmp1-2 mat+ strain grown at 30° compared to the wild-type and Δoxa1 (oxa1ts) rmp1-1 mat− strains grown under the same conditions. It appears to be equivalent to that observed in the complex IV-deficient long-lived mutant cox5:: ble, which strengthens the idea of a strong link between life span and ROS production in P. anserina.
DISCUSSION
In this study, we have identified the oxa1 gene of P. anserina. It is functionally homologous to the S. cerevisiae oxa1 gene and is essential for viability in P. anserina. A strain carrying a thermosensitive copy of the gene (oxa1ts), grown at subrestrictive temperature, exhibits severe defects in the respiratory complexes I and IV. One of the most striking results obtained in this study is the existence of an interaction between genes oxa1 and rmp1. The effects of complex I and IV activity impairment are an increased life span, a strong induction of the alternative oxidase, and a decrease in ROS production. Results presented here show that there is no causal link between the increase of life span and the amount of alternative oxidase.
The P. anserina OXA1 protein is functionally homologous to the S. cerevisiae protein and is essential for viability:
The P. anserina OXA1 protein is able to complement the respiratory defect of a S. cerevisiae null mutant, as was previously shown for the OXA1 proteins from S. pombe (Bonnefoy et al. 2000), N. crassa (Nargang et al. 2002), Arabidopsis thaliana (Hamel et al. 1997), and H. sapiens (Bonnefoy et al. 1994b), demonstrating that all these proteins are truly functional homologs. In S. cerevisiae, a single transition mutation that results in an L240S substitution in a hydrophilic region between the second and third transmembrane domains leads to a respiratory thermosensitive phenotype (Bauer et al. 1994; Meyer et al. 1997b). The Leu240 residue is conserved in the different OXA1 homologs, ranging from bacteria to human (Meyer et al. 1997b). We show here that a change of the corresponding leucine to serine (L269S) in the P. anserina protein also leads to a thermosensitive phenotype, supporting the view that this conserved residue is essential for the function of OXA1 proteins.
The absence of Oxa1p in S. cerevisiae and that of the Sp2 ortholog in S. pombe lead to a complete loss of respiration. However, the knockout of the oxa1 gene in S. cerevisiae is viable (Bonnefoy et al. 1994a), whereas the knockout of the two S. pombe orthologs, Sp1 and Sp2, is nonviable (Bonnefoy et al. 2000). It was proposed that this lethality is linked to the small negative phenomenon and that the loss of Oxa1p function leads to severe defects in the F1-ATPase, resulting in the incapacity to ensure a basal level of membrane potential (Bonnefoy et al. 2000). The OXA1 function is of course expected to be essential for viability in strictly aerobic organisms, in which all the electron flow is transported through the cytochrome pathway. In organisms possessing an alternative respiration system branched to the cytochrome pathway, such as the filamentous fungi, the question of the OXA1 function requirement deserved to be raised. However, in both N. crassa and P. anserina the absence of OXA1 was shown to be nonviable. The lethality of Δoxa1 mutants cannot be explained by defects in complex IV biogenesis since loss of function of cytochrome c oxidase is viable in P. anserina. It can therefore be concluded that it is due to complex V defects as hypothesized for S. pombe and/or to complex I defects. This complex, absent in S. cerevisiae and S. pombe, is situated upstream of the branch point of the alternative oxidase and therefore cannot be compensated by it. In N. crassa, depletion of OXA1 leads to reduced levels of the 24- and 29.9-kD subunits of complex I revealed in Western blot (Nargang et al. 2002). We show here that in a rmp1-2 context, a compromised OXA1 activity reduces complex I activity dramatically and, to a lesser extent, complex IV activity, whereas it has practically no effect on complex V activity.
Genetic interaction between the oxa1 and rmp1 genes:
A remarkable and exciting result provided by this work is the occurrence of a genetic interaction between the oxa1 and rmp1 genes. The two natural alleles rmp1-1 and rmp1-2 differ by three substitutions but the life-span differences observed between AS1-4 rmp1-1 and AS1-4 rmp1-2 have been attributed to a premature stop, which yields a protein lacking its last 19 amino acids in the rmp1-2 context. Several lines of evidence support the hypothesis that rmp1-1 is the fully functional allele (Contamine et al. 2004). Interestingly, the defects in the biogenesis of complexes I and IV in Δoxa1 (oxa1ts) grown at 30° are much more pronounced in a rmp1-2 than in a rmp1-1 context, suggesting that the mutant form of OXA1 displays dependency on the RMP1 functional protein to play its role in the biogenesis of the respiratory complexes. The synergic relationship between the mutant OXA1 and the supposed not fully functional form of RMP1 suggests that the two encoded proteins are involved in the same process. The function of RMP1 is unknown and putative homologs have been found only in multicellular ascomycetes (Contamine et al. 2004). However, the authors hypothesize that the RMP1 function could be widely distributed and could have evolved so rapidly that recognition of its homologs in distant species would be impaired. These authors have reported database searches indicating that the Histoplasma capsulatum homolog to RMP1 presents a putative homolog in S. pombe, which itself shows a weak similarity to the S. cerevisiae Sls1p protein. Sls1p is a mitochondrial membrane protein required for respiration (Rouillard et al. 1996). Its precise function remains to be elucidated. Recent data indicate that Sls1p is involved in mitochondrial translation regulation in cooperation with Nam1p and could serve to coordinate transcription and translation of mtDNA encoded products (Bryan et al. 2002; Rodeheffer and Shadel 2003). It was also recently reported that Oxa1p may serve as a ribosome-binding site that allows coupling of the synthesis and membrane integration of mtDNA encoded proteins (Jia et al. 2003; Szyrach et al. 2003). The two proteins could therefore interact at least during some cellular stages. Furthermore, on the basis of the fact that overexpression of SLS1 does not rescue the translation defect of mutants of the amino terminal domain of the mtRNA polymerase but increases the steady-state level of some mitochondrial encoded proteins, it has been proposed that Sls1p could be involved in the assembly of the oxidative phosphorylation complexes in a manner independent of its effects on translation (Rodeheffer and Shadel 2003). Regardless the significance of these observations, the genetic interaction between the oxa1 and rmp1 genes revealed in this study points to a role for the RMP1 protein in the process of OXA1-dependent protein insertion and provides a new framework for further studies.
Impairment of complex I and IV activities leads to increased life span and strong induction of the alternative oxidase but these two parameters are not causally linked:
This study confirms the influence of the mitochondrial activity in the control of life span and is consistent with previous work using respiratory-deficient mutants (Dufour et al. 2000; Lorin et al. 2001; Stumpferl et al. 2004). In the Δoxa1 (oxa1ts) rmp1-1 strains grown at subrestrictive temperature, the defects in complexes I and IV are nonexistent or modest, and life span is slightly reduced. In the Δoxa1 (oxa1ts) rmp1-2 strains, in contrast, complexes I and IV appear strongly affected, and life span is considerably extended. Defects in complexes I and IV activities in rmp1-2 strains are paralleled with a strong induction of the alternative oxidase, a shift of the partitioning of the electrons through this oxidase, leading to a highly SHAM-sensitive respiration and to a decrease in ROS production (Dufour et al. 2000; Lorin et al. 2001; Stumpferl et al. 2004). If there is no doubt that close links occur between life span, AOX level, and ROS production in respiratory-deficient mutants of P. anserina, the nature of these links is still obscure. A correlation between increased life span and induced aox expression has been recently reported (Borghouts et al. 2002; Stumpferl et al. 2004). On the other hand, it was also shown that a high level of AOX due to the constitutive overexpression of the alternative oxidase in long-lived respiration mutants results in shortening life span and in restoring senescence (Lorin et al. 2001). We show here that neither the overexpression of the alternative oxidase in the short-lived Δoxa1 (oxa1ts) rmp1-1 strain nor the inactivation of the aox resident gene in the long-lived Δoxa1 (oxa1ts) rmp1-2 strain significantly modifies the life span of these strains. This result clearly establishes that there is no causal relationship between the AOX level and life span.
The physiological role of the alternative pathway is still puzzling. It is of course important under conditions in which the cytochrome pathway is impaired. However, expression of the alternative oxidase can rescue mitochondrial respiration only if complex I is functional since the branch point is located downstream of this complex. We describe here for the first time a mutant displaying a defect in complex I activity in P. anserina. This defect is associated with a defect in complex IV and with a strong induction of the AOX, which, however, does not seem to play a crucial role in the control of life span since inactivation of the aox gene has no apparent effect. The induction of the AOX in a complex I-deficient mutant is, a priori, surprising and could only be a general response to a mitochondrial dysfunction, independent of its “usefulness” (which of course is evident for loss-of-function mutants of the cytochrome pathway). Although it was reported that ROS could induce the expression of the alternative oxidase in plants (Wagner 1995; Yukioka et al. 1998), the precise nature of the signal(s) that leads to this induction is unknown to date.
We previously showed that inactivation of complex IV in P. anserina results in an increase of life span associated with a decrease in the levels of free radicals (Dufour et al. 2000). Interestingly, the data obtained in this study also show that in strains mutated for OXA1, life span increase is correlated with ROS decrease. An impaired respiratory chain often increases oxidative stress (Ishii et al. 1998; McLennan and Degli Esposti 2000; Harman 2001). The reverse effect observed in the Δcox5 mutant of P. anserina was attributed to the presence of an alternative oxidase that could play a protective role in mitochondria by preventing ROS production. However, to date, this hypothesis has not been clearly demonstrated in P. anserina (Lorin et al. 2001). The reason(s) why deficiency in complex I in the Δoxa1 (oxa1ts) rmp1-2 strain is associated with a reduction in ROS level is not yet well understood. Importantly, the presented data confirm the correlation between life-span increase and ROS decrease, supporting the idea that ROS level is a leading cause of aging.
Future studies will focus on deciphering the nature of the links between ROS production, AOX induction, and control of life span. They will also focus on the characterization of the interaction between the genes oxa1 and rmp1. This characterization may hold a key for understanding the role of RMP1 in the control of mtDNA stability and for identification of its potential functional homologs in other species.
Footnotes
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. AF382189.
Communicating editor: P. J. Pukkila
Acknowledgement
We thank M. Picard and L. Sperling for helpful discussions and comments on the manuscript. We gratefully acknowledge M. Minet and V. Rincheval for their help in the library construction and flow cytometric analysis, respectively. We also warmly thank V. Contamine for the gift of pHSS plasmid and T. Elthon and J. Velours for their gift of antibodies against AOX and β-subunit of ATPase, respectively. We are grateful to C. Barreau for providing chromoslot membranes and to M. Kermorgant and A. Léon for their help in the plasmid construction. This work was supported by grants from the Association Française contre les Myopathies.
References
Albert, B., and C. H. Sellem,
Altamura, N., N. Capitanio, N. Bonnefoy, S. Papa and G. Dujardin,
Barreau, C., C. Sellem, P. Silar, A. Sainsard-Chanet and B. Turcq,
Bauer, M., M. Behrens, K. Esser, G. Michaelis and E. Pratje,
Belcour, L., O. Begel and M. Picard,
Belcour, L., A. Sainsard-Chanet, C. Jamet-Vierny and M. Picard, 1999 Stability of the mitochondrial genome of Podospora anserina and its genetic control, pp. 209–228 in Mitochondrial Disease, edited by P. Lestienne. Springer-Verlag, Berlin/New York
Berges, T., and C. Barreau,
Bonnefoy, N., F. Chalvet, P. Hamel, P. P. Slonimski and G. Dujardin,
Bonnefoy, N., M. Kermorgant, O. Groudinsky, M. Minet, P. P. Slonimski et al.,
Bonnefoy, N., M. Kermorgant, O. Groudinsky and G. Dujardin,
Borghouts, C., A. Werner, T. Elthon and H. D. Osiewacz,
Borghouts, C., C. Q. Scheckhuber, A. Werner and H. D. Osiewacz,
Bryan, A. C., M. S. Rodeheffer, C. M. Wearn and G. S. Shadel,
Contamine, V., G. Lecellier, L. Belcour and M. Picard,
Contamine, V., D. Zickler and M. Picard,
Coppin, E., and R. Debuchy,
Debuchy, R., S. Arnaise and G. Lecellier,
d'Enfert, C., M. Minet and F. Lacroute,
Dequard-Chablat, M., and C. H. Sellem,
Dequard-Chablat, M., E. Coppin-Raynal, M. Picard-Bennoun and J. J. Madjar,
Dillin, A., A. L. Hsu, N. Arantes-Oliveira, J. Lehrer-Graiwer, H. Hsin et al.,
Dufour, E., J. Boulay, V. Rincheval and A. Sainsard-Chanet,
Elthon, T., R. L. Nickels and L. McIntosh,
Esser, K., 1974 Podospora anserina, pp. 531–551 in Handbook of Genetics, edited by R. C. King, Plenum Press, New York.
Feng, J., F. Bussiere and S. Hekimi,
Hamel, P., W. Sakamoto, H. Wintz and G. Dujardin,
Hamel, P., C. Lemaire, N. Bonnefoy, P. Brivet-Chevillotte and G. Dujardin,
Harman, D.,
Harman, P.,
Herrmann, J. M., and W. Neupert,
Ishii, N., M. Fujii, P. S. Hartman, M. Tsuda, K. Yasuda et al.,
Jia, L., M. Dienhart, M. Schramp, M. McCauley, K. Hell et al.,
Kuhn, A., R. Stuart, R. Henry and R. E. Dalbey,
Lee, S. S., R. Y. Lee, A. G. Fraser, R. S. Kamath, J. Ahringer et al.,
Lorin, S., E. Dufour, J. Boulay, O. Begel, S. Marsy et al.,
McLennan, H. R., and M. Degli Esposti,
Meyer, W., M. Bauer and E. Pratje,
Meyer, W., U. Bomer and E. Pratje,
Munkres, K.,
Nargang, F. E., M. Preuss, W. Neupert and J. M. Herrmann,
Nijtmans, L. G., N. S. Henderson and I. J. Holt,
Rizet, G.,
Rizet, G.,
Rizet, G., and C. Engelmann,
Rodeheffer, M. S., and G. S. Shadel,
Rouillard, J. M., M. E. Dufour, B. Theunissen, E. Mandart, G. Dujardin et al.,
Schagger, H., and G. von Jagow,
Sellem, C. H., A. Sainsard-Chanet and L. Belcour,
Shigenaga, M. K., T. M. Hagen and B. N. Ames,
Sluse, F. E., and W. Jarmuszkiewicz,
Stumpferl, S. W., O. Stephan and H. D. Osiewacz,
Szyrach, G., M. Ott, N. Bonnefoy, W. Neupert and J. M. Herrmann,
Trifunovic, A., A. Wredenberg, M. Falkenberg, J. N. Spelbrink, A. T. Rovio et al.,
Turcq, B., 1989 Clonage direct de gènes par complementation chez le champignon Podospora anserina: application à l'étude de gènes d'incompatibilité. Ph.D. Thesis, University of Bordeaux II, Bordeaux, France.
Vanlerberghe, G. C., and L. McIntosh,
Wagner, A. M.,
Yukioka, H., S. Inagaki, R. Tanaka, K. Katoh, N. Miki et al.,