-
PDF
- Split View
-
Views
-
Cite
Cite
James M Mason, Joshua Ransom, Alexander Y Konev, A Deficiency Screen for Dominant Suppressors of Telomeric Silencing in Drosophila, Genetics, Volume 168, Issue 3, 1 November 2004, Pages 1353–1370, https://doi.org/10.1534/genetics.104.030676
Close - Share Icon Share
Abstract
Heterochromatin is a specialized chromatin structure in chromosomal regions associated with repeated DNA sequences and low concentrations of genes. Formation of heterochromatin is determined in large part by enzymes that modify histones and structural proteins that bind to these modified histones in a cooperative fashion. In Drosophila, mutations in genes that encode heterochromatic proteins are often dominant and increase expression of genes placed into heterochromatic positions. To find components of telomeric heterochromatin in Drosophila, we screened a collection of autosomal deficiencies for dominant suppressors of silencing of a transgene at the telomere of chromosome 2L. While many deficiency chromosomes are associated with dominant suppressors, in the cases tested on chromosome 2 the suppressor mapped to the 2L telomere, rather than the deficiency. We infer that background effects may hamper the search for genes that play a role in telomeric heterochromatin formation and that either very few genes participate in this pathway or mutations in these genes are not dominant suppressors of telomeric position effect. The data also suggest that the 2L telomere region plays a major role in telomeric silencing.
TELOMERES are structures at the ends of linear chromosomes that are required for chromosome stability. They allow the linear DNA molecules to complete the replication of chromosome ends. Telomeres also cap chromosome ends, which would otherwise resemble DNA double-strand breaks. In addition, telomeres form a domain of transcriptionally repressed chromatin.
A prominent characteristic of telomeres is heterochromatin-like organization of surrounding chromatin. Silencing [termed telomeric position effect (TPE)] is observed when genes are placed near telomeres in Trypanosoma brucei (Horn and Cross 1995; Rudenkoet al. 1995), Saccharomyces cerevisiae (Gottschlinget al. 1990; Palladino and Gasser 1994), Schizosaccharomyces pombe (Nimmoet al. 1994), Drosophila melanogaster (Gehringet al. 1984; Hazelrigget al. 1984; Karpen and Spradling 1992; Leviset al. 1993; Wallrath and Elgin 1995), and humans (Bauret al. 2001). Such a widespread conservation of telomeric silencing among eukaryotes suggests that it is fundamental to telomere function. Indeed, telomere length maintenance and TPE in yeast appear to be tightly connected (Kyrionet al. 1993; Parket al. 2002).
Telomeres typically consist of a tandem array of GC-rich telomeric DNA repeats specified by copying of the template sequence within the telomerase RNA. These DNA repeats bind a set of sequence-specific DNA-binding proteins that, through separate domains, bind additional proteins to assemble an inferred higher-order complex nucleated on the telomeric DNA repeats (Blackburn 2001). In S. cerevisiae, where TPE is most extensively studied (Dubranaet al. 2001; Shore 2001), Rap1p binds to multiple sites within the telomeric repeats and, together with chromosome end-binding proteins yKu70p and yKu80p, recruits the silent information regulation silencing complex (Kyrionet al. 1993; Morettiet al. 1994; Boulton and Jackson 1998; Larocheet al. 1998).
Chromosome ends in D. melanogaster do not terminate in an array of simple repeats that is synthesized by telomerase, as in other species. Instead, Drosophila uses two families of non-long terminal repeat retrotransposons, HeT-A and TART, to elongate its chromosome ends (Mason and Biessmann 1995). Proximal to the terminal retrotransposon array Drosophila telomeres carry several kilobases of complex satellites, termed telomere-associated sequences (TAS), which exhibit sequence similarities among themselves (Karpen and Spradling 1992; Walteret al. 1995) and structural similarities to TAS in other eukaryotes (Prydeet al. 1997). Despite the fact that Drosophila does not possess arrays of simple repeats, such as those that bind Rap1p, Drosophila shares the property of telomeric silencing with other organisms. Reporter genes exhibit repressed and variegated expression when inserted into Drosophila telomeric regions (Gehringet al. 1984; Hazelrigget al. 1984; Leviset al. 1985; Karpen and Spradling 1992; Toweret al. 1993; Rosemanet al. 1995; Wallrath and Elgin 1995; Masonet al. 2000, 2003a). As all variegating telomeric transgenes analyzed to date are embedded in or lie adjacent to TAS (Karpen and Spradling 1992; Leviset al. 1993; Wallrath and Elgin 1995; Crydermanet al. 1999; Marinet al. 2000; Golubovskyet al. 2001), TAS appears to play a role in telomeric silencing. This was demonstrated directly using a transgenic approach (Kurenovaet al. 1998). In Drosophila, variegated repression of telomeric transgenes resembles position-effect variegation (PEV), the clonal inactivation of a euchromatic gene that has been positioned close to or within centric heterochromatin (Weiler and Wakimoto 1995). TPE, however, appears to be qualitatively different from PEV, because genetic modifiers of PEV, including the presence of an extra Y chromosome, have no effect on repression of transgenes inserted into TAS sequences (Talbertet al. 1994; Wallrath and Elgin 1995; Crydermanet al. 1999). Many suppressors of PEV in Drosophila are found to encode components of the repressive multimeric protein complex bound to centric heterochromatin or enzymes involved in their modification (Weiler and Wakimoto 1995).
Another well-known example of genetic silencing in Drosophila, developmentally regulated gene silencing, is mediated by proteins encoded by the Polycomb group (PcG) of genes (Pirrotta 1995). Many mutations in PcG genes, however, do not affect TPE. Exceptions are the weak suppression by certain alleles of Psc, Su(z)2, and possibly a few other loci, and the stronger suppression by the small deficiency, Su(z)25, which deletes Su(z)2, and Psc (Crydermanet al. 1999; Boivinet al. 2003). Thus, while a repressive chromatin complex is likely formed at Drosophila telomeres, components of this complex remain unknown.
We recently characterized the molecular structure of P{wvar}, a variegating insertion of a genomic white gene in the 2L telomere (Gehringet al. 1984; Golubovskyet al. 2001). Unlike other repressed telomeric reporter genes, the transgene in P{wvar} is located precisely between the terminal retrotransposon array and TAS (Golubovskyet al. 2001). P{wvar} is very sensitive to its context; changes in the structure of the telomere region, such as HeT-A additions to the chromosome terminus, terminal deficiencies, gradual loss from the chromosome end due to incomplete replication, and loss of the 2L TAS region on the homolog, can be identified easily by changes in eye color. Considering the sensitivity of P{wvar} to changes at the 2L telomere in cis as well as in trans, we reasoned that this insertion and its derivatives might be a sensitive model for selection of trans-modifiers of TPE.
Here we report the results of a screen for dominant trans-acting TPE modifiers on autosomal deficiency chromosomes maintained at the Bloomington Drosophila stock center (http://flystocks.bio.indiana.edu/). While many of the second chromosomes tested carried suppressors of TPE, in every case examined in detail the suppressor mapped to the 2L tip, rather than to the site of the deficiency. In addition, several of these chromosomes fail to hybridize a 2L TAS probe in situ, and some fail to complement lethal mutations at l(2)gl, a gene very close to the 2L telomere. While the third chromosome deficiencies were not characterized in detail, these results indicate that genetic background effects may be a serious complication when analyzing the ability of extant mutants to suppress TPE. They also confirm reports (Golubovskyet al. 2001) that deficiencies of the 2L telomere strongly suppress silencing of a reporter gene in the homologous tips.
MATERIALS AND METHODS
Drosophila crosses:
Drosophila stocks were maintained and crosses were performed at 25° on cornmeal, molasses medium with dry yeast added to the surface. The y1 w67c23; P{wvar} stock has been described recently (Golubovskyet al. 2001), and P{wvar}KR3-2 is a stable “brown-red” variant of P{wvar}. Other genetic markers and special chromosomes are described by Lindsley and Zimm (1992) and/or FlyBase (FlyBaseConsortium 2003). Su(z)25 was kindly supplied by L. L. Wallrath, and Psc1 was a generous gift of S. Ronsseray.
Suppression of telomeric silencing:
Deficiency (Df) chromosomes obtained from the Bloomington stock center were tested for suppression of TPE by crossing y w67c23; P{wvar}KR3-2 females to Df/Balancer males and scoring the eye color of y w67c23; P{wvar}KR3-2 sons with and without the Balancer. Males with a light orange eye color were designated nonsuppressor. Males with darker eye color were considered to carry a suppressor. At least five males of each genotype were examined before a determination was made. If the eye color of Df males overlapped the color of Balancer males, a more careful comparison was made. At least six Df and at least six Balancer males, 8–48 hr old, were arranged in order according to eye color, and the Mann-Whitney rank order test was used to identify suppressors using the tables in Mendenhall (1971).
If Balancer males had dark eyes, Df/Balancer males were crossed with y w67c23; Balancer females (Sco/SM1 for chromosome 2 deficiencies; Sb/TM6 for chromosome 3 deficiencies), and the deficiency chromosome was retested for a suppressor phenotype. In some cases repeated backcrosses to y w67c23; Balancer females were required to get a consistent result. To identify a suppressor on the Df chromosome the resulting Balancer males were required to have orange eyes, while Df males had dark eyes. That is, the suppressor must segregate with the Deficiency chromosome to be considered. In the first test for suppression of TPE, a parallel cross of y w67c23 females to Df/Balancer males was made to control for the presence of cryptic white insertions on the deficiency chromosomes.
Ambiguities in the reported deficiency breakpoints made the identification of putative sites of dominant suppressors problematic. For purposes of constructing a map, we made the assumption that the deficiencies on a chromosome with a suppressor phenotype include the region of ambiguity, but the nonsuppressor deficiencies do not. This overestimates the number and extent of putative suppressor sites. Breakpoints reported by FlyBase (FlyBaseConsortium 2003) are used here, as these positions are determined by genetic as well as cytological data. The proportion of the genome uncovered by the deficiencies tested was estimated by counting euchromatic bands; i.e., regions 40, 41, 80, and 81 were not counted.
Lethal complementation tests:
Complementation tests were made by crossing l(2)gl/SM1 females to Df/Cy balancer males in small mass matings. After 4 days parents were transferred to a second vial. F1 progeny were counted through day 17 or until at least two straight-winged flies were recovered. Thus, viability is defined operationally as two adult test flies. Crosses lacking straight-winged progeny were repeated until there were at least 60 Cy progeny or, if the two Cy classes could be distinguished, until at least 20 of each class emerged. Independent tests were made with two alleles of l(2)gl in different genetic backgrounds, l(2)gl26 and l(2)glM1. The latter is a terminal deficiency for 2L TAS that acquired a lethal allele of l(2)gl by terminal erosion (A. Y. Konev and J. M. Mason, unpublished results). A deficiency chromosome must fail to complement both alleles to be considered to carry a mutation for l(2)gl.
Recombination mapping:
Second chromosome deficiencies were combined with y w67c23 by crossing deficiency males to y w67c23; Sco/SM1 females and then mating Sco+ Cy F1 brothers and sisters and selecting for y and w progeny in the F2.
A stock of al S wgSp-1 Tft nwB PinYt/CyO was obtained from the Bloomington stock center and males from this stock were crossed to y w67c23 females to replace the X chromosome. As the multiply marked chromosome had poor viability in the presence of y w67c23 and Cy, the line was maintained by backcrossing y w67c23; + females to y w67c23; al S wgSp-1 Tft nwB PinYt/+ males each generation. Only second chromosome modifiers were mapped by recombination, because multiply marked third chromosomes from the stock center carried dominant suppressors of telomeric silencing.
For mapping studies, y w67c23; Df/SM1 females were crossed to y w67c23; al S wgSp-1 Tft nwB PinYt/+ males. To map lethal mutations on the deficiency chromosome, the F1 Cy+ multiply marked females were backcrossed to y w67c23; Df/SM1 males, and F2 Cy+ progeny were scored for the dominant visible markers. To map the suppressors on the deficiency chromosome, the F1 Cy+ multiply marked females were crossed to y w67c23; P{wvar}KR3-2 al males, and progeny were scored for eye color as well as the other visible chromosome 2 markers. Test crosses consisted of small mass matings. As the P{wvar}KR3-2 chromosome carries al, but the deficiency chromosomes do not, al could be used as a marker for mapping the suppressors, but not the lethals. At least 100 chromosomes were counted to map the suppressors, and 100 Cy+ chromosomes were counted to map the lethals.
In situ hybridization:
Salivary chromosome squashes of larvae from deficiency stocks were prepared according to Kurenova et al. (1998). The balancer breakpoints were used as cytological markers to identify the 2L telomere region. A 6-kb fragment of the 2L TAS array (Kurenovaet al. 1998) was used as probe, and the 2L TAS array on the balancer acted as a hybridization control. To confirm that hybridization occurred with the balancer rather than with the deficiency chromosome, several deficiencies were retested from a y w67c23; Df/SM1 stock, where the SM1 balancer chromosome is known to hybridize strongly to the 2L TAS probe.
RESULTS
A screen for suppression of telomeric silencing:
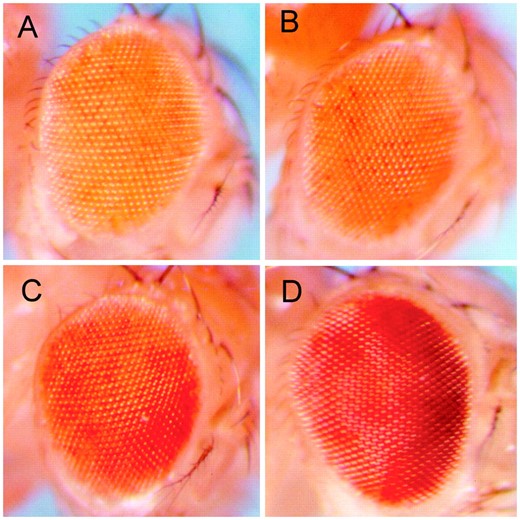
To inquire into the existence of suppressors of TPE in Drosophila and simultaneously map their positions, we screened the autosomal deficiency kits from the Bloomington stock center for a suppressor phenotype. y w67c23; P{wvar}KR3-2 females were mated to Df/Balancer males and the eye color of y w67c23 males with and without the Balancer was determined. Males with a light orange eye color were considered to lack a suppressor of TPE; males with darker eye color were considered to carry a suppressor (Figure 1
Eye color phenotypes of suppressors of telomeric silencing. All photographs show hemizygous P{wvar}KR3-2. Equivocal suppressors, labeled “+” in Table 1, have a phenotype that overlaps the nonsuppressed phenotype and are not shown here. (A) The absence of a suppressor of TPE, labeled “−” in Table 1. (B) A weak suppressor of TPE, labeled ++ in Table 1. (C) A moderate suppressor of TPE, labeled +++. (D) A strong suppressor of TPE, labeled ++++.
; Table 1)
Deficiency chromosomes tested for suppression of telomeric silencing
Df no. . | Breakpointsa . | Name . | Suppressionb . | Callc . |
|---|---|---|---|---|
| 1 | 21A1; 21B6–7 | Df(2L)net-PM47C | ++ | FP4 |
| 2 | 21A1; 21B7–8 | Df(2L)net-PMF | + | FP4, -5 |
| 3 | 21A1–4; 21B3 | Df(2L)net18 | ++ | FP4, -5 |
| 4 | 21A1–4; 21B4 | Df(2L)net62 | ++ | FP4, -5 |
| 5 | 21C1; 21C7 | Df(2L)al | ++ | Region 1 |
| 6 | 21C3–4; 21C6–8 | Df(2L)BSC16 | − | |
| 7 | 21C8–D1; 22A8–B1 | Df(2L)S2 | +++ | FP4 |
| 8 | 21D1–2; 21E1–2 | Df(2L)ast4 | +++ | FP4, -5 |
| 9 | 21D1–2; 22B2–3 | Df(2L)ast2 | + | FP6 |
| 10 | 21D2; 21F3–22A1 | Df(2L)S3 | + | FP6 |
| 11 | 21E3–4; 22B5–7 | Df(2L)frtz17 | − | |
| 12 | 22A1; 22B6–9 | D(2L)frtz11 | ++ | FP1 |
| 13 | 22A2–3; 22B7 | D(2L)frtz19 | ++ | FP1 |
| 14 | 22A2–3; 22D5–E1 | Df(2L)dp-79b | − | |
| 15 | 22A3; 22B3 | Df(2L)frtz14 | + | FP1, -6 |
| 16 | 22A3–4; 22C1–2 | Df(2L)frtz25 | +++ | FP1 |
| 17 | 22A6; 22B9 | Df(2L)J69LH56R | ++ | FP1 |
| 18 | 22F1–2; 23A2 | Df(2L)dpp-d14 | − | |
| 19 | 22F3–4; 23C3–5 | Df(2L)C144 | − | |
| 20 | 22F4; 23A1 | Df(2L)D20 | ++ | FP1, -4 |
| 21 | 23A3–4; 23D4–6 | Df(2L)JS13 | ++ | Region 2 |
| 22 | 23C1–2; 23E1–2 | Df(2L)JS17 | ++ | Region 2 |
| 23 | 23C3–5; 23D1–2 | Df(2L)JS32 | − | |
| 24 | 23D2; 23E3 | Df(2L)S2590 | ++ | FP2, -4 |
| 25 | 23F2–3; 23F6–24A1 | Df(2L)tim-02 | − | |
| 26 | 24A2; 24D4 | Df(2L)ed1 | − | |
| 27 | 24C3; 25A2 | Df(2L)ed-dp | + | FP6 |
| 28 | 24E1; 25A2 | Df(2L)M24F-B | − | |
| 29 | 24E3; 25A7 | Df(2L)sc19-3 | − | |
| 30 | 24E4; 25B2 | Df(2L)dp-h25 | − | |
| 31 | 24F1–2; 25C5 | Df(2L)sc19-6 | − | |
| 32 | 25A5; 25D6 | Df(2L)sc19-5 | ++ | Region 3 |
| 33 | 25D2–3; 26B2–5 | Df(2L)cl-h3 | − | |
| 34 | 25F3–26A1; 26D3–11 | Df(2L)E110 | − | |
| 35 | 26D3–E1; 26F4–7 | Df(2L)BSC6 | − | |
| 36 | 26D10–E1; 27C1 | Df(2L)BSC7 | ++ | Region 4 |
| 37 | 27B2; 27F1–2 | Df(2L)spd-j2 | − | |
| 38 | 27C5–9; 28B3–4 | Df(2L)J-H | − | |
| 39 | 27E; 28C1–4 | Df(2L)spd | + | FP6 |
| 40 | 27E2; 28D1 | Df(2L)XE-3801 | +++ | FP4 |
| 41 | 28B2; 28D1 | Df(2L)XE-2750 | + | FP6 |
| 42 | 28DE (within) | Df(2L)Trf-C6R31 | ++ | FP4 |
| 43 | 28E4–7; 29B2–C1 | Df(2L)TE29Aa-11 | − | |
| 44 | 29C1–2; 30C8–9 | Df(2L)N22-14 | ++ | FP4 |
| 45 | 29C3–5; 30C8–9 | Df(2L)N22-5 | ++ | Regions 5, 6 |
| 46 | 29E2–F1; 30C2–4 | Df(2L)TE30Cb-1 | − | |
| 47 | 30A1–2; 30D1–2 | Df(2L)N22-3 | ++ | Region 6 |
| 48 | 30A3–5; 30C5 | Df(2L)30A-C | ++ | Region 6 |
| 49 | 30A9–B1; 30D2–F4 | Df(2L)gamma7 | +++ | FP4 |
| 50 | 30D1–F6; 31F1–5 | Df(2L)Mdh | − | |
| 51 | 31B1; 32A1–2 | Df(2L)J2 | − | |
| 52 | 31D1–11; 31E7 | Df(2L)J27 | − | |
| 53 | 32D1; 32F1–3 | Df(2L)FCK-20 | − | |
| 54 | 32F1–3; 33F1–2 | Df(2L)Prl | + | FP5, -6 |
| 55 | 33A1; 33B1–2 | Df(2L)esc-P2-0 | + | FP6 |
| 56 | 33A1; 33B2 | Df(2L)esc10 | − | |
| 57 | 33A1; 33E | Df(2L)esc-P3-0 | − | |
| 58 | 33B3; 34A1–2 | Df(2L)prd1.7 | − | |
| 59 | 34B7–12; 34E3 | In(2L)b82a1 | ++ | Region 7 |
| 60 | 34C1; 35C1 | Df(2L)b87e25 | ++ | FP2, -5 |
| 61 | 34C4; 35A4 | Df(2L)b80e3 | − | |
| 62 | 34D1–2; 35C1 | Df(2L)64j | +++ | FP1 |
| 63 | 34D3–4; 35C1 | Df(2L)fn30 | ++++ | FP1 |
| 64 | 34D4; 34E3 | Df(2L)b88f32 | ++ | FP1 |
| 65 | 34E2; 35B3–4 | Df(2L)fn7 | +++ | FP1 |
| 66 | 34E3; 35D2–5 | Df(2L)el80f1 | + | FP1, -6 |
| 67 | 34E4–34F1; 35C3 | Df(2L)noc11 | − | |
| 68 | 34E5–F1; 35C3 | Df(2L)A263 | − | |
| 69 | 34F2–5; 35C4 | Df(2L)osp38 | ++ | FP1 |
| 70 | 34F4; 35C3 | Df(2L)fn5 | − | |
| 71 | 34F4–5; 35D4–5 | Df(2L)fn1 | ++++ | FP1 |
| 72 | 34F5; 35B2 | Df(2L)el81i1 | − | |
| 73 | 34F5; 35B10 | Df(2L)TE35BC-31 | − | |
| 74 | 34F5–35A4; 35D2 | Df(2L)do1 | − | |
| 75 | 35A1–4; 35C1–3 | Df(2L)A400 | − | |
| 76 | 35A4–B1; 35B2 | Df(2L)fn2 | + | FP1 |
| 77 | 35B1; 35F1 | Df(2L)A446 | − | |
| 78 | 35B3; 35E6 | Df(2L)osp29 | − | |
| 79 | 35B4–6; 35E1–2 | Df(2L)TE35BC-24 | − | |
| 80 | 35D1; 36A6–7 | Df(2L)r10 | ++ | Region 8 |
| 81 | 35F6–12; 36D | Df(2L)cact-255rv64 | +++ | FP5 |
| 82 | 36A8–9; 36F1 | Df(2L)H20 | − | |
| 83 | 36C2–4; 37B9–10 | Df(2L)TW137 | − | |
| 84 | 36E4–F1; 38A6–7 | Df(2L)TW50 | +++ | FP1 |
| 85 | 36F7–9; 37B2–7 | Df(2L)TW3 | − | |
| 86 | 36F7–9; 37D1–2 | Df(2L)VA16 | ++ | FP1 |
| 87 | 37B2–8; 37C5 | Df(2L)hk-UC2 | − | |
| 88 | 37B2–10; 38D2–5 | Df(2L)pr-A16 | − | |
| 89 | 37B2–8; 37E2 | Df(2L)TW158 | ++ | FP1 |
| 90 | 37B9–10; 37D1–2 | Df(2L)TW130 | ++ | FP1 |
| 91 | 37B9–10; 37D5 | Df(2L)VA23 | ++ | FP1 |
| 92 | 37C1; 37F5 | Df(2L)VA17 | ++ | FP1 |
| 93 | 37C2–5; 38B2–C1 | Df(2L)VA12 | +++ | FP1 |
| 94 | 37C2–7; 38C1–2 | Df(2L)Sd77 | − | |
| 95 | 37D2; 38A1 | Df(2L)E55 | +++ | FP1 |
| 96 | 37D2–5; 38A6–B2 | Df(2L)Sd37 | − | |
| 97 | 37D2–5; 39A4–7 | Df(2L)pr-A14 | ++ | FP1 |
| 98 | 37D6–E1; 38E6–9 | Df(2L)TW2 | ++ | FP1 |
| 99 | 37E2–4; 39D1 | Df(2L)TW12 | − | |
| 100 | 37E2–F1; 38B5–C1 | Df(2L)TW9 | +++ | FP1 |
| 101 | 38A1; 39D3–E1 | Df(2L)TW84 | + | FP1, -6 |
| 102 | 38A1; 39F1 | Df(2L)TW65 | ++ | Region 9 |
| 103 | 38A3–4; 38B6–C1 | Df(2L)pr-A20 | ++ | FP1 |
| 104 | 38A7–B1; 39C2–3 | Df(2L)TW1 | ++ | FP1 |
| 105 | 38B3–6; 40A3 | Df(2L)pr-M1 | + | FP6 |
| 106 | 38E2; 39E7 | Df(2L)DS6 | − | |
| 107 | 40h35; 40h38L | Df(2L)C' | − | |
| 108 | h38R; h46 | Df(2R)M41A10 | + | FP6 |
| 109 | h42–h43; 42A2–3 | In(2R)bwVDe2LCyR | + | FP6 |
| 110 | h44–h46; 41B1–41F11 | Df(2R)M41A8 | +++ | FP4, -5 |
| 111 | h44–h46; 42A1–2 | Df(2R)M41A4 | ++ | FP4 |
| 112 | 41BC; 42A16–B1 | Df(2R)nap14 | + | FP6 |
| 113 | 41D2–E1; 42B1–3 | Df(2R)nap1 | − | |
| 114 | 41F3–4; 42A3–9 | Df(2R)17I | ++ | FP1 |
| 115 | 42A1–2; 42E6–F1 | Df(2R)nap9 | − | |
| 116 | 42A1–19; 42E2–7 | Df(2R)cn88b | − | |
| 117 | 42B3–4; 43E18 | Df(2R)ST1 | ++ | FP1 |
| 118 | 42B4–C1; 43F–44A1 | Df(2R)cn87e | − | |
| 119 | 42C1–7; 43F5–8 | Df(2R)pk78s | − | |
| 120 | 42C2; 42D2–3 | Df(2R)42 | − | |
| 121 | 42C2–7; 43D1–7 | Df(2R)Drl-rv17 | + | FP1, -6 |
| 122 | 42E; 44C1 | Df(2R)cn9 | ++ | FP1 |
| 123 | 42E1–4; 43C3 | Df(2R)Drl-rv3 | ++ | FP1 |
| 124 | 43A3; 43F6 | Df(2R)P32a | + | FP1, -6 |
| 125 | 43C5; 44B6–C1 | Df(2R)cn83c | + | FP1, -6 |
| 126 | 43C7; 43F2–8 | Df(2R)cn-S6 | − | |
| 127 | 43E7–18; 44B4–5 | Df(2R)CA53 | − | |
| 128 | 43F; 44D3–8 | Df(2R)H3C1 | − | |
| 129 | 44C1–2; 44E1–4 | Df(2R)44CE | − | |
| 130 | 44D1–4; 44F12 | Df(2R)H3E1 | − | |
| 131 | 44D2–E1; 45B8–C1 | Df(2R)Np3 | − | |
| 132 | 44F11; 45C1 | Df(2R)Np4 | ++ | FP1 |
| 133 | 44F11; 45D9–E1 | Df(2R)Np5 | + | FP1, -6 |
| 134 | 44F2–3; 45C6 | Df(2R)Np1 | − | |
| 135 | 45A6–7; 45E2–3 | Df(2R)w45-30n | + | FP4 |
| 136 | 45A9–10; 45D5–8 | Df(2R)w73-1 | − | |
| 137 | 45C8; 45D8 | Df(2R)wun-GL | ++ | FP1 |
| 138 | 45C8–D10; 45D9–E1 | Df(2R)w45-19g | ++ | FP1 |
| 139 | 45D3–4; 45F2–6 | Df(2R)BSC29 | − | |
| 140 | 46A1–4; 46C3–12 | Df(2R)B5 | + | FP6 |
| 141 | 46C1–2; 46E1–2 | Df(2R)X3 | − | |
| 142 | 46C2; 47A1 | Df(2R)X1 | − | |
| 143 | 46C3–4; 46C9–11 | Df(2R)eve | +++ | FP1, -2 |
| 144 | 46F1; 47A10 | Df(2R)12 | ++ | FP2 |
| 145 | 46F1; 47B9 | Df(2R)stan2 | ++ | FP3 |
| 146 | 47A3; 47E | Df(2R)E3363 | − | |
| 147 | 47D3; 48A5 | Df(2R)en-A | + | FP6 |
| 148 | 47E3; 48A5–B2 | Df(2R)en-B | ++ | FP2, -4 |
| 149 | 48A1; 48B5 | Df(2R)en-SFX31 | +++ | FP3 |
| 150 | 48A1–2; 48B–C1 | Df(2R)en28 | + | FP3, -6 |
| 151 | 48A3; 48C6–8 | Df(2R)en30 | − | |
| 152 | 48C5–D1; 48D5–E11 | Df(2R)BSC39 | − | |
| 153 | 48E; 49A | Df(2R)CB21 | +++ | FP2, -5 |
| 154 | 49A; 49E1–2 | Df(2R)vg135 | + | FP5, -6 |
| 155 | 49B2–3; 49E2 | Df(2R)vg-C | − | |
| 156 | 49C1–2; 49E6 | Df(2R)vg-D | ++ | Region 10 |
| 157 | 49C1–4; 50C23–D1 | Df(2R)CX1 | +++ | FP4 |
| 158 | 49D3–4; 50A2 | Df(2R)vg-B | + | FP6 |
| 159 | 50C21–23; 50D1–5 | Df(2R)50C-101 | − | |
| 160 | 50E6–F1; 51E2–4 | Df(2R)BSC11 | +++ | FP4 |
| 161 | 50F6–9; 51B3 | Df(2R)L48 | − | |
| 162 | 51A2; 51B6 | Df(2R)trix | ++ | FP1 |
| 163 | 51A5; 51C1 | Df(2R)03072 | − | |
| 164 | 51C3–7; 51E7–11 | Df(2R)14 | − | |
| 165 | 51D1–2; 51E5 | Df(2R)XTE-58 | − | |
| 166 | 51D3–E1; 52D1 | Df(2R)XTE-18 | − | |
| 167 | 51F13; 52F8–9 | Df(2R)Jp4 | +++ | Region 11 |
| 168 | 52A13–14; 52F10–11 | Df(2R)Jp5 | +++ | Region 11 |
| 169 | 52A9–10; 52D9–15 | Df(2R)WMG | − | |
| 170 | 52D3; 53A1 | Df(2R)Jp6 | ++ | Region 11 |
| 171 | 54E8–F1; 55B9–C1 | Df(2R)Pcl7B | − | |
| 172 | 54F2; 56A1 | Df(2R)RM2-1 | +++ | FP3 |
| 173 | 55A1; 55C1–3 | Df(2R)Pcl11B | − | |
| 174 | 55A1; 55F1–2 | Df(2R)PC4 | − | |
| 175 | 55C1–2; 56B1–2 | Df(2R)C29 | ++ | FP3 |
| 176 | 55D2–E1; 56B2 | Df(2R)PC66 | ++ | FP3 |
| 177 | 55E6–F3; 56C1 | Df(2R)P34 | ++ | FP2 |
| 178 | 56D7–E3; 56F9–12 | Df(2R)BSC22 | + | FP6 |
| 179 | 56F 5; 56F15 | Df(2R)173 | − | |
| 180 | 56F 5; 56F15 | Df(2R)017 | − | |
| 181 | 56F 9–11; 57D12 | Df(2R)AA21 | + | FP3, -6 |
| 182 | 56F12–14; 57A4 | Df(2R)BSC19 | − | |
| 183 | 57A3; 57B1 | Df(2R)exu2 | +++ | FP2 |
| 184 | 57A6; 57B6 | Df(2R)D4 | − | |
| 185 | 57B1; 57B13–14 | Df(2R)E2 | − | |
| 186 | 57B4; 58B1–2 | Df(2R)Pu-D17 | − | |
| 187 | 57D2–8; 58D1 | Df(2R)Egfr5 | − | |
| 188 | 58B1–2; 58E4 | Df(2R)X58-7 | − | |
| 189 | 58B3; 59A1 | Df(2R)X58-8 | − | |
| 190 | 58C3–7; 58D6–8 | Df(2R)X58-3 | − | |
| 191 | 58D1–2; 59A1 | Df(2R)X58-12 | +++ | Region 12 |
| 192 | 59A1–3; 59B1–2 | Df(2R)59AB | ++ | Region 12 |
| 193 | 59A1–3; 59D1–4 | Df(2R)59AD | − | |
| 194 | 59D 4–8; 59D9–E1 | Df(2R)vir-12 | − | |
| 195 | 59D 8; 60A7 | Df(2R)bw-S46 | + | FP6 |
| 196 | 59D11; 59F6–8 | Df(2R)bw-HB132 | + | FP4 |
| 197 | 59E; 60A1 | Df(2R)egl2 | ++ | FP1 |
| 198 | 59E1; 59F6 | Df(2R)bw5 | + | FP6 |
| 199 | 59E1; 60C7–D1 | Df(2R)bwVDe2LPxKR | − | |
| 200 | 59F1; 59F5 | Df(2R)egl3 | ++ | FP1 |
| 201 | 59F3; 60A8–16 | Df(2R)G10-7-5 | − | |
| 202 | 59F6; 60A12–16 | Df(2R)or-BR11 | − | |
| 203 | 60B8–10; 60D1 | Df(2R)Px1 | − | |
| 204 | 60C6; 60D9–10 | Df(2R)Px2 | − | |
| 205 | 60E6; 60F1–2 | Df(2R)ES1 | ++ | FP2, -4 |
| 206 | 60E1–2; 60E6 | Df(2R)Dll-MP | ++ | FP4 |
| 207 | 60E10; 60F5 | Df(2R)Kr10 | ++ | FP4 |
| 208 | 60E6–9; 60E11 | Df(2R)M60E | − | |
| 209 | 60E9; 60F1 | Df(2R)gsb | + | FP3, -6 |
| 210 | 60F2; 60F5 | Df(2R)Kr14 | − | |
| 211 | 61A; 61D3 | Df(3L)emc-E12 | − | |
| 212 | 61A1; 61B | Df(3L)B71 | − | |
| 213 | 61C1–4; 61F3 | Df(3L)Ar12-1 | + | FP6 |
| 214 | 61C3–4; 61E | Df(3L)Ar11 | + | FP6 |
| 215 | 61C4; 62A8 | Df(3L)Ar14-8 | + | FP6 |
| 216 | 61D3–E1; 61F5–8 | Df(3L)bab-PG | − | |
| 217 | 61F8; 62A3–5 | Df(3L)ru-22 | + | FP6 |
| 218 | 62A10–B1; 62C4–D1 | Df(3L)R-G5 | − | |
| 219 | 62A10–B1; 62D2 | Df(3L)Aprt-1 | − | |
| 220 | 62B9; 62E7 | Df(3L)R-G7 | − | |
| 221 | 63C1; 63D3 | Df(3L)HR232 | − | |
| 222 | 63C1–2; 63F1–2 | Df(3L)1227 | ++ | FP1 |
| 223 | 63C6; 63F7 | Df(3L)HR119 | − | |
| 224 | 63E2; 64B17 | Df(3L)GN50 | − | |
| 225 | 63F6–7; 64C13–15 | Df(3L)GN24 | − | |
| 226 | 64; 65B5–C1 | Dr(3L)CH39 | − | |
| 227 | 64B–C; 65B5–C1 | Df(3L)CH18 | − | |
| 228 | 64C; 65C | Df(3L)ZN47 | − | |
| 229 | 64E1–13; 65C1–D6 | Df(3L)v65c | ++ | FP1 |
| 230 | 65A; 65E1 | Df(3L)W5.4 | − | |
| 231 | 65A2; 65E1 | Df(3L)XDl98 | − | |
| 232 | 65D4–5; 65E4–6 | Df(3L)BSC27 | − | |
| 233 | 65E1–12; 66A17 | Df(3L)RM5-2 | − | |
| 234 | 65F3; 66B10 | Df(3L)pbl-X1 | − | |
| 235 | 66A17–20; 66C1–5 | Df(3L)ZP1 | − | |
| 236 | 66B12–C1; 66D2–4 | Df(3L)BSC13 | ++ | Region 13 |
| 237 | 66B8–9; 66C9–10 | Df(3L)66C-G28 | +++ | Region 13 |
| 238 | 66C7–10; 66C7–10 | Df(3L)66C-I65 | − | |
| 239 | 66E1–6; 66F1–6 | Df(3L)Scf-R6 | − | |
| 240 | 66E3–4; 66F1–2 | Df(3L)Scf-R11 | − | |
| 241 | 66F5; 66F5 | Df(3L)Rdl-2 | − | |
| 242 | 66F5; 67B1 | Df(3L)29A6 | ++ | Region 14 |
| 243 | 67A2; 67D13 | Df(3L)AC1 | + | FP6 |
| 244 | 67E1–2; 68C1–2 | Df(3L)lxd6 | − | |
| 245 | 68A2; 69A1 | Df(3L)vin5 | − | |
| 246 | 68C8; 69B4–5 | Df(3L)vin7 | − | |
| 247 | 69A4–5; 69D4–6 | Df(3L)eyg-C1 | − | |
| 248 | 69B1–5; 69D1–6 | Df(3L)iro-2 | − | |
| 249 | 69D; 69D | Df(3L)8ex25 | + | FP1, -6 |
| 250 | 69D2; 69E3–5 | Df(3L)E44 | − | |
| 251 | 69D4–5; 69F5–7 | Df(3L)BSC10 | − | |
| 252 | 69F3–4; 70C3–4 | Df(3L)C190LUbx42TR | − | |
| 253 | 70C2; 72A1 | Df(3L)D-5rv12 | − | |
| 254 | 70C2–6; 70E1 | Df(3L)fz-CAL | − | |
| 255 | 70D2; 70E8 | Df(3L)fz-D21 | − | |
| 256 | 70D2; 71E4–5 | Df(3L)fz-M21 | − | |
| 257 | 70E; 71F | Df(3L)Brd6 | ++ | FP1 |
| 258 | 71A1–2; 71C1–2 | Df(3L)Brd15 | − | |
| 259 | 71C2–3; 72B1–C1 | Df(3L)XG-5 | − | |
| 260 | 71C3; 71E5 | Df(3L)BK10 | − | |
| 261 | 72A2; 72D10 | Df(3L)th102 | − | |
| 262 | 72A3–4; 72D1–5 | Df(3L)brm11 | − | |
| 263 | 72C1; 73A4 | Df(3L)st-f13 | − | |
| 264 | 72D10–11; 73D1–2 | Df(3L)st-b11 | − | |
| 265 | 73A3; 74F1–4 | Df(3L)81k19 | − | |
| 266 | 74D3–75A1; 75B2–5 | Df(3L)BSC8 | − | |
| 267 | 75B10; 75C5 | Df(3L)W4 | − | |
| 268 | 75C1–2; 75F1 | Df(3L)Cat | − | |
| 269 | 75F10–11; 76A1–5 | Df(3L)fz2 | − | |
| 270 | 76A7–B1; 76B4–5 | Df(3L)BSC20 | − | |
| 271 | 76B; 76F | Df(3L)XS2182 | − | |
| 272 | 76B; 77A | Df(3L)XS543 | − | |
| 273 | 76B1–2; 76D5 | Df(3L)kto2 | ++ | FP1 |
| 274 | 76B4; 76D3 | Df(3L)XS705 | − | |
| 275 | 76B4; 77B | Df(3L)XS-533 | − | |
| 276 | 76B6; 77C1 | Df(3L)XS572 | − | |
| 277 | 77A1; 77D1 | Df(3L)rdgC-co2 | − | |
| 278 | 77B7–9; 77F1–5 | Df(3L)ri-79c | ++ | Region 15 |
| 279 | 77E2; 78A4 | Df(3L)ri-XT1 | − | |
| 280 | 77F3; 78C8–9 | Df(3L)ME107 | − | |
| 281 | 78C5–6; 78E3–79A1 | Df(3L)Pc-2q | − | |
| 282 | 79C; 79E1–8 | Df(3L)Ten-m-AL29 | − | |
| 283 | 79D3–E1; 79F3–6 | Df(3L)HD1 | − | |
| 284 | 79E1–2; 79E1–8 | Df(3L)Ten-m-AL1 | − | |
| 285 | 79E5–F1; 80A2–3 | Df(3L)BSC21 | − | |
| 286 | 79F; 80A | Df(3L)Delta1AK | − | |
| 287 | 80Fb–g | Df(3L)3-52 | + | FP6 |
| 288 | 80Fd–e | Df(3R)6-61 | + | FP6 |
| 289 | 80Ff–g | Df(3L)8A-80 | − | |
| 290 | 80Fg–j | Df(3L)1-166 | − | |
| 291 | 80Fh–j | Df(3L)2-66 | + | FP6 |
| 292 | 80Fj | Df(3L)2-30 | + | FP6 |
| 293 | 81 Fab | Df(3R)4-75 | − | |
| 294 | 81F; 82F10–11 | Df(3R)2-2 | − | |
| 295 | 81F3–6; 82F5–7 | Df(3R)ME15 | + | FP6 |
| 296 | 82A5–6; 82E4 | Df(3R)Z1 | − | |
| 297 | 82C4; 82F3 | Df(3R)110 | − | |
| 298 | 82D5; 82F3–6 | Df(3R)6-7 | + | FP1, -6 |
| 299 | 82F3–4; 82F10–11 | Df(3R)3-4 | − | |
| 300 | 82F8–10; 83A1–3 | Df(3R)e1025-14 | − | |
| 301 | 83B7–C1; 83C6–D1 | Df(3R)BSC47 | − | |
| 302 | 83E1–2; 84B1 | Df(3R)WIN11 | ++ | Region 16 |
| 303 | 83E3; 84B1 | Df(3R)Dfd13 | +++ | Region 16 |
| 304 | 84A1; 84B1 | Df(3R)9A99 | − | |
| 305 | 84A1–2; 84B1–2 | Df(3R)Scr | − | |
| 306 | 84A6–B1; 84D4–D9 | Df(3R)roe | − | |
| 307 | 84C1–2; 84E1 | Df(3R)dsx2M | − | |
| 308 | 84C8; 84F6 | Df(3R)dsx29 | − | |
| 309 | 84D 4–6; 85B6 | Df(3R)p712 | ++ | FP1 |
| 310 | 84D 8; 85B3–5 | Df(3R)dsx37 | − | |
| 311 | 84D 8–9; 85A1–2 | Df(3R)dsx11 | − | |
| 312 | 84D11; 84E8 | Df(3R)dsx15 | − | |
| 313 | 84E8–9; 85B6 | Df(3R)p40 | ++ | FP1 |
| 314 | 84F1; 85A6–B9 | Df(3R)p13 | − | |
| 315 | 84F2; 85A5–7 | Df(3R)CA3 | ++ | FP1 |
| 316 | 85A2; 85C1–2 | Df(3R)p-XT103 | − | |
| 317 | 85D8; 85E10–13 | Df(3R)by10 | − | |
| 318 | 85D11–13; 85F6 | Df(3R)by62 | +++ | Region 17 |
| 319 | 85D12; 85E10 | Df(3R)GB104 | − | |
| 320 | 86C1; 87B5 | Df(3R)M-Kx1 | − | |
| 321 | 86E2–3; 87C6–7 | Df(3R)T-32 | − | |
| 322 | 86F1–2; 87C6–7 | Df(3R)T-10 | ++ | FP1 |
| 323 | 87B12; 87E8 | Df(3R)ry615 | − | |
| 324 | 87D2; 87F2 | Df(3R)ry27 | − | |
| 325 | 87E–F; 88B | Df(3R)CbxTwtLUbxKM5R | + | FP1, -6 |
| 326 | 87E1; 87F11 | Df(3R)I26c | − | |
| 327 | 87F1; 87F15 | Df(3R)urd | − | |
| 328 | 87F12–14; 88C2 | Df(3R)red31 | − | |
| 329 | 88A2; 88C1–D1 | Df(3R)red1 | − | |
| 330 | 88B1; 88C2 | Df(3R)red-P93 | − | |
| 331 | 88E7–13; 89A1 | Df(3R)ea | + | FP6 |
| 332 | 88F7; 89A11–13 | Df(3R)Po4 | − | |
| 333 | 89A1–2; 89A11–13 | Df(3R)Po2 | ++ | FP1 |
| 334 | 89A1–2; 89A11–13 | Df(3R)Po3 | − | |
| 335 | 89A8; 89B3 | Df(3R)Exel7327 | − | |
| 336 | 89B4; 89B10 | Df(3R)sbd45 | − | |
| 337 | 89B5; 89C | Dr(3R)sd104 | − | |
| 338 | 89B5–6; 89E2–3 | Df(3R)bxd100 | + | FP1, -6 |
| 339 | 89B7–8; 89E7–8 | Df(3R)P115 | − | |
| 340 | 89E1–89F4; 91B1–B2 | Df(3R)DG2 | − | |
| 341 | 89E2–3; 90A | Df(3R)C4 | ++ | FP1 |
| 342 | 89E2–3; 90D | Df(3R)RD31 | − | |
| 343 | 90F1–2; 91F5 | Df(3R)Cha7 | − | |
| 344 | 91A2–B3; 91F13–92A1 | Df(3R)Cha1a | − | |
| 345 | 91F1–2; 92D3–6 | Df(3R)D1-BX12 | − | |
| 346 | 92B3; 92F13 | Df(3R)H-B79 | − | |
| 347 | 93B2–13; 94A3–8 | Df(3R)e-N19 | − | |
| 348 | 93B6; 93D3–4 | Df(3R)e-R1 | − | |
| 349 | 93C6; 94A1–4 | Df(3R)e-GC3 | + | FP1, -6 |
| 350 | 93E–F; 94C–D | Df(3R)5C1 | + | FP1, -6 |
| 351 | 93F11–14; 94D10–13 | Df(3R)hh | − | |
| 352 | 95A5–7; 95C10–11 | Df(3R)mbc-30 | ++ | Region 18 |
| 353 | 95A5–7; 95D6–11 | Df(3R)mbc-R1 | − | |
| 354 | 95D7–11; 95F15 | Df(3R)crb-F89-4 | − | |
| 355 | 95D11–E2; 96A2 | Df(3R)crb87-4 | − | |
| 356 | 96A1; 96A20–25 | Df(3R)Ubx7LLatsR | − | |
| 357 | 96A2–7; 96D2–4 | Df(3R)slo8 | − | |
| 358 | 96F1; 97B1 | Df(3R)Espl3 | − | |
| 359 | 96F10–11; 96F11 | Df(3R)Espl22 | − | |
| 360 | 96F12–14; 97C4–5 | Df(3R)ME61 | − | |
| 361 | 97A; 98A1–2 | Df(3R)T1-P | − | |
| 362 | 97E3; 98A5 | Df(3R)D605 | − | |
| 363 | 98D3–7; 98D3–7 | Df(3R)M15 | − | |
| 364 | 98E3; 99A6 | Df(3R)3450 | − | |
| 365 | 98F14; 99E2–3 | Df(3R)R133 | − | |
| 366 | 99A6; 99C1 | Df(3R)01215 | − | |
| 367 | 99D1–2; 99E1 | Df(3R)X3F | − | |
| 368 | 99F1–2; 100B5 | Df(3R)tll-g | − | |
| 369 | 100A2; 100C2–3 | Df(3R)tll-e | + | FP6 |
| 370 | 100D1; 100D3–4 | Df(3R)awd-KRB | − | |
| 371 | 100D1–2; 100E–F | Df(3R)faf-BP | − | |
| 372 | 100D2; 100F5 | Df(3R)04661 | − |
Df no. . | Breakpointsa . | Name . | Suppressionb . | Callc . |
|---|---|---|---|---|
| 1 | 21A1; 21B6–7 | Df(2L)net-PM47C | ++ | FP4 |
| 2 | 21A1; 21B7–8 | Df(2L)net-PMF | + | FP4, -5 |
| 3 | 21A1–4; 21B3 | Df(2L)net18 | ++ | FP4, -5 |
| 4 | 21A1–4; 21B4 | Df(2L)net62 | ++ | FP4, -5 |
| 5 | 21C1; 21C7 | Df(2L)al | ++ | Region 1 |
| 6 | 21C3–4; 21C6–8 | Df(2L)BSC16 | − | |
| 7 | 21C8–D1; 22A8–B1 | Df(2L)S2 | +++ | FP4 |
| 8 | 21D1–2; 21E1–2 | Df(2L)ast4 | +++ | FP4, -5 |
| 9 | 21D1–2; 22B2–3 | Df(2L)ast2 | + | FP6 |
| 10 | 21D2; 21F3–22A1 | Df(2L)S3 | + | FP6 |
| 11 | 21E3–4; 22B5–7 | Df(2L)frtz17 | − | |
| 12 | 22A1; 22B6–9 | D(2L)frtz11 | ++ | FP1 |
| 13 | 22A2–3; 22B7 | D(2L)frtz19 | ++ | FP1 |
| 14 | 22A2–3; 22D5–E1 | Df(2L)dp-79b | − | |
| 15 | 22A3; 22B3 | Df(2L)frtz14 | + | FP1, -6 |
| 16 | 22A3–4; 22C1–2 | Df(2L)frtz25 | +++ | FP1 |
| 17 | 22A6; 22B9 | Df(2L)J69LH56R | ++ | FP1 |
| 18 | 22F1–2; 23A2 | Df(2L)dpp-d14 | − | |
| 19 | 22F3–4; 23C3–5 | Df(2L)C144 | − | |
| 20 | 22F4; 23A1 | Df(2L)D20 | ++ | FP1, -4 |
| 21 | 23A3–4; 23D4–6 | Df(2L)JS13 | ++ | Region 2 |
| 22 | 23C1–2; 23E1–2 | Df(2L)JS17 | ++ | Region 2 |
| 23 | 23C3–5; 23D1–2 | Df(2L)JS32 | − | |
| 24 | 23D2; 23E3 | Df(2L)S2590 | ++ | FP2, -4 |
| 25 | 23F2–3; 23F6–24A1 | Df(2L)tim-02 | − | |
| 26 | 24A2; 24D4 | Df(2L)ed1 | − | |
| 27 | 24C3; 25A2 | Df(2L)ed-dp | + | FP6 |
| 28 | 24E1; 25A2 | Df(2L)M24F-B | − | |
| 29 | 24E3; 25A7 | Df(2L)sc19-3 | − | |
| 30 | 24E4; 25B2 | Df(2L)dp-h25 | − | |
| 31 | 24F1–2; 25C5 | Df(2L)sc19-6 | − | |
| 32 | 25A5; 25D6 | Df(2L)sc19-5 | ++ | Region 3 |
| 33 | 25D2–3; 26B2–5 | Df(2L)cl-h3 | − | |
| 34 | 25F3–26A1; 26D3–11 | Df(2L)E110 | − | |
| 35 | 26D3–E1; 26F4–7 | Df(2L)BSC6 | − | |
| 36 | 26D10–E1; 27C1 | Df(2L)BSC7 | ++ | Region 4 |
| 37 | 27B2; 27F1–2 | Df(2L)spd-j2 | − | |
| 38 | 27C5–9; 28B3–4 | Df(2L)J-H | − | |
| 39 | 27E; 28C1–4 | Df(2L)spd | + | FP6 |
| 40 | 27E2; 28D1 | Df(2L)XE-3801 | +++ | FP4 |
| 41 | 28B2; 28D1 | Df(2L)XE-2750 | + | FP6 |
| 42 | 28DE (within) | Df(2L)Trf-C6R31 | ++ | FP4 |
| 43 | 28E4–7; 29B2–C1 | Df(2L)TE29Aa-11 | − | |
| 44 | 29C1–2; 30C8–9 | Df(2L)N22-14 | ++ | FP4 |
| 45 | 29C3–5; 30C8–9 | Df(2L)N22-5 | ++ | Regions 5, 6 |
| 46 | 29E2–F1; 30C2–4 | Df(2L)TE30Cb-1 | − | |
| 47 | 30A1–2; 30D1–2 | Df(2L)N22-3 | ++ | Region 6 |
| 48 | 30A3–5; 30C5 | Df(2L)30A-C | ++ | Region 6 |
| 49 | 30A9–B1; 30D2–F4 | Df(2L)gamma7 | +++ | FP4 |
| 50 | 30D1–F6; 31F1–5 | Df(2L)Mdh | − | |
| 51 | 31B1; 32A1–2 | Df(2L)J2 | − | |
| 52 | 31D1–11; 31E7 | Df(2L)J27 | − | |
| 53 | 32D1; 32F1–3 | Df(2L)FCK-20 | − | |
| 54 | 32F1–3; 33F1–2 | Df(2L)Prl | + | FP5, -6 |
| 55 | 33A1; 33B1–2 | Df(2L)esc-P2-0 | + | FP6 |
| 56 | 33A1; 33B2 | Df(2L)esc10 | − | |
| 57 | 33A1; 33E | Df(2L)esc-P3-0 | − | |
| 58 | 33B3; 34A1–2 | Df(2L)prd1.7 | − | |
| 59 | 34B7–12; 34E3 | In(2L)b82a1 | ++ | Region 7 |
| 60 | 34C1; 35C1 | Df(2L)b87e25 | ++ | FP2, -5 |
| 61 | 34C4; 35A4 | Df(2L)b80e3 | − | |
| 62 | 34D1–2; 35C1 | Df(2L)64j | +++ | FP1 |
| 63 | 34D3–4; 35C1 | Df(2L)fn30 | ++++ | FP1 |
| 64 | 34D4; 34E3 | Df(2L)b88f32 | ++ | FP1 |
| 65 | 34E2; 35B3–4 | Df(2L)fn7 | +++ | FP1 |
| 66 | 34E3; 35D2–5 | Df(2L)el80f1 | + | FP1, -6 |
| 67 | 34E4–34F1; 35C3 | Df(2L)noc11 | − | |
| 68 | 34E5–F1; 35C3 | Df(2L)A263 | − | |
| 69 | 34F2–5; 35C4 | Df(2L)osp38 | ++ | FP1 |
| 70 | 34F4; 35C3 | Df(2L)fn5 | − | |
| 71 | 34F4–5; 35D4–5 | Df(2L)fn1 | ++++ | FP1 |
| 72 | 34F5; 35B2 | Df(2L)el81i1 | − | |
| 73 | 34F5; 35B10 | Df(2L)TE35BC-31 | − | |
| 74 | 34F5–35A4; 35D2 | Df(2L)do1 | − | |
| 75 | 35A1–4; 35C1–3 | Df(2L)A400 | − | |
| 76 | 35A4–B1; 35B2 | Df(2L)fn2 | + | FP1 |
| 77 | 35B1; 35F1 | Df(2L)A446 | − | |
| 78 | 35B3; 35E6 | Df(2L)osp29 | − | |
| 79 | 35B4–6; 35E1–2 | Df(2L)TE35BC-24 | − | |
| 80 | 35D1; 36A6–7 | Df(2L)r10 | ++ | Region 8 |
| 81 | 35F6–12; 36D | Df(2L)cact-255rv64 | +++ | FP5 |
| 82 | 36A8–9; 36F1 | Df(2L)H20 | − | |
| 83 | 36C2–4; 37B9–10 | Df(2L)TW137 | − | |
| 84 | 36E4–F1; 38A6–7 | Df(2L)TW50 | +++ | FP1 |
| 85 | 36F7–9; 37B2–7 | Df(2L)TW3 | − | |
| 86 | 36F7–9; 37D1–2 | Df(2L)VA16 | ++ | FP1 |
| 87 | 37B2–8; 37C5 | Df(2L)hk-UC2 | − | |
| 88 | 37B2–10; 38D2–5 | Df(2L)pr-A16 | − | |
| 89 | 37B2–8; 37E2 | Df(2L)TW158 | ++ | FP1 |
| 90 | 37B9–10; 37D1–2 | Df(2L)TW130 | ++ | FP1 |
| 91 | 37B9–10; 37D5 | Df(2L)VA23 | ++ | FP1 |
| 92 | 37C1; 37F5 | Df(2L)VA17 | ++ | FP1 |
| 93 | 37C2–5; 38B2–C1 | Df(2L)VA12 | +++ | FP1 |
| 94 | 37C2–7; 38C1–2 | Df(2L)Sd77 | − | |
| 95 | 37D2; 38A1 | Df(2L)E55 | +++ | FP1 |
| 96 | 37D2–5; 38A6–B2 | Df(2L)Sd37 | − | |
| 97 | 37D2–5; 39A4–7 | Df(2L)pr-A14 | ++ | FP1 |
| 98 | 37D6–E1; 38E6–9 | Df(2L)TW2 | ++ | FP1 |
| 99 | 37E2–4; 39D1 | Df(2L)TW12 | − | |
| 100 | 37E2–F1; 38B5–C1 | Df(2L)TW9 | +++ | FP1 |
| 101 | 38A1; 39D3–E1 | Df(2L)TW84 | + | FP1, -6 |
| 102 | 38A1; 39F1 | Df(2L)TW65 | ++ | Region 9 |
| 103 | 38A3–4; 38B6–C1 | Df(2L)pr-A20 | ++ | FP1 |
| 104 | 38A7–B1; 39C2–3 | Df(2L)TW1 | ++ | FP1 |
| 105 | 38B3–6; 40A3 | Df(2L)pr-M1 | + | FP6 |
| 106 | 38E2; 39E7 | Df(2L)DS6 | − | |
| 107 | 40h35; 40h38L | Df(2L)C' | − | |
| 108 | h38R; h46 | Df(2R)M41A10 | + | FP6 |
| 109 | h42–h43; 42A2–3 | In(2R)bwVDe2LCyR | + | FP6 |
| 110 | h44–h46; 41B1–41F11 | Df(2R)M41A8 | +++ | FP4, -5 |
| 111 | h44–h46; 42A1–2 | Df(2R)M41A4 | ++ | FP4 |
| 112 | 41BC; 42A16–B1 | Df(2R)nap14 | + | FP6 |
| 113 | 41D2–E1; 42B1–3 | Df(2R)nap1 | − | |
| 114 | 41F3–4; 42A3–9 | Df(2R)17I | ++ | FP1 |
| 115 | 42A1–2; 42E6–F1 | Df(2R)nap9 | − | |
| 116 | 42A1–19; 42E2–7 | Df(2R)cn88b | − | |
| 117 | 42B3–4; 43E18 | Df(2R)ST1 | ++ | FP1 |
| 118 | 42B4–C1; 43F–44A1 | Df(2R)cn87e | − | |
| 119 | 42C1–7; 43F5–8 | Df(2R)pk78s | − | |
| 120 | 42C2; 42D2–3 | Df(2R)42 | − | |
| 121 | 42C2–7; 43D1–7 | Df(2R)Drl-rv17 | + | FP1, -6 |
| 122 | 42E; 44C1 | Df(2R)cn9 | ++ | FP1 |
| 123 | 42E1–4; 43C3 | Df(2R)Drl-rv3 | ++ | FP1 |
| 124 | 43A3; 43F6 | Df(2R)P32a | + | FP1, -6 |
| 125 | 43C5; 44B6–C1 | Df(2R)cn83c | + | FP1, -6 |
| 126 | 43C7; 43F2–8 | Df(2R)cn-S6 | − | |
| 127 | 43E7–18; 44B4–5 | Df(2R)CA53 | − | |
| 128 | 43F; 44D3–8 | Df(2R)H3C1 | − | |
| 129 | 44C1–2; 44E1–4 | Df(2R)44CE | − | |
| 130 | 44D1–4; 44F12 | Df(2R)H3E1 | − | |
| 131 | 44D2–E1; 45B8–C1 | Df(2R)Np3 | − | |
| 132 | 44F11; 45C1 | Df(2R)Np4 | ++ | FP1 |
| 133 | 44F11; 45D9–E1 | Df(2R)Np5 | + | FP1, -6 |
| 134 | 44F2–3; 45C6 | Df(2R)Np1 | − | |
| 135 | 45A6–7; 45E2–3 | Df(2R)w45-30n | + | FP4 |
| 136 | 45A9–10; 45D5–8 | Df(2R)w73-1 | − | |
| 137 | 45C8; 45D8 | Df(2R)wun-GL | ++ | FP1 |
| 138 | 45C8–D10; 45D9–E1 | Df(2R)w45-19g | ++ | FP1 |
| 139 | 45D3–4; 45F2–6 | Df(2R)BSC29 | − | |
| 140 | 46A1–4; 46C3–12 | Df(2R)B5 | + | FP6 |
| 141 | 46C1–2; 46E1–2 | Df(2R)X3 | − | |
| 142 | 46C2; 47A1 | Df(2R)X1 | − | |
| 143 | 46C3–4; 46C9–11 | Df(2R)eve | +++ | FP1, -2 |
| 144 | 46F1; 47A10 | Df(2R)12 | ++ | FP2 |
| 145 | 46F1; 47B9 | Df(2R)stan2 | ++ | FP3 |
| 146 | 47A3; 47E | Df(2R)E3363 | − | |
| 147 | 47D3; 48A5 | Df(2R)en-A | + | FP6 |
| 148 | 47E3; 48A5–B2 | Df(2R)en-B | ++ | FP2, -4 |
| 149 | 48A1; 48B5 | Df(2R)en-SFX31 | +++ | FP3 |
| 150 | 48A1–2; 48B–C1 | Df(2R)en28 | + | FP3, -6 |
| 151 | 48A3; 48C6–8 | Df(2R)en30 | − | |
| 152 | 48C5–D1; 48D5–E11 | Df(2R)BSC39 | − | |
| 153 | 48E; 49A | Df(2R)CB21 | +++ | FP2, -5 |
| 154 | 49A; 49E1–2 | Df(2R)vg135 | + | FP5, -6 |
| 155 | 49B2–3; 49E2 | Df(2R)vg-C | − | |
| 156 | 49C1–2; 49E6 | Df(2R)vg-D | ++ | Region 10 |
| 157 | 49C1–4; 50C23–D1 | Df(2R)CX1 | +++ | FP4 |
| 158 | 49D3–4; 50A2 | Df(2R)vg-B | + | FP6 |
| 159 | 50C21–23; 50D1–5 | Df(2R)50C-101 | − | |
| 160 | 50E6–F1; 51E2–4 | Df(2R)BSC11 | +++ | FP4 |
| 161 | 50F6–9; 51B3 | Df(2R)L48 | − | |
| 162 | 51A2; 51B6 | Df(2R)trix | ++ | FP1 |
| 163 | 51A5; 51C1 | Df(2R)03072 | − | |
| 164 | 51C3–7; 51E7–11 | Df(2R)14 | − | |
| 165 | 51D1–2; 51E5 | Df(2R)XTE-58 | − | |
| 166 | 51D3–E1; 52D1 | Df(2R)XTE-18 | − | |
| 167 | 51F13; 52F8–9 | Df(2R)Jp4 | +++ | Region 11 |
| 168 | 52A13–14; 52F10–11 | Df(2R)Jp5 | +++ | Region 11 |
| 169 | 52A9–10; 52D9–15 | Df(2R)WMG | − | |
| 170 | 52D3; 53A1 | Df(2R)Jp6 | ++ | Region 11 |
| 171 | 54E8–F1; 55B9–C1 | Df(2R)Pcl7B | − | |
| 172 | 54F2; 56A1 | Df(2R)RM2-1 | +++ | FP3 |
| 173 | 55A1; 55C1–3 | Df(2R)Pcl11B | − | |
| 174 | 55A1; 55F1–2 | Df(2R)PC4 | − | |
| 175 | 55C1–2; 56B1–2 | Df(2R)C29 | ++ | FP3 |
| 176 | 55D2–E1; 56B2 | Df(2R)PC66 | ++ | FP3 |
| 177 | 55E6–F3; 56C1 | Df(2R)P34 | ++ | FP2 |
| 178 | 56D7–E3; 56F9–12 | Df(2R)BSC22 | + | FP6 |
| 179 | 56F 5; 56F15 | Df(2R)173 | − | |
| 180 | 56F 5; 56F15 | Df(2R)017 | − | |
| 181 | 56F 9–11; 57D12 | Df(2R)AA21 | + | FP3, -6 |
| 182 | 56F12–14; 57A4 | Df(2R)BSC19 | − | |
| 183 | 57A3; 57B1 | Df(2R)exu2 | +++ | FP2 |
| 184 | 57A6; 57B6 | Df(2R)D4 | − | |
| 185 | 57B1; 57B13–14 | Df(2R)E2 | − | |
| 186 | 57B4; 58B1–2 | Df(2R)Pu-D17 | − | |
| 187 | 57D2–8; 58D1 | Df(2R)Egfr5 | − | |
| 188 | 58B1–2; 58E4 | Df(2R)X58-7 | − | |
| 189 | 58B3; 59A1 | Df(2R)X58-8 | − | |
| 190 | 58C3–7; 58D6–8 | Df(2R)X58-3 | − | |
| 191 | 58D1–2; 59A1 | Df(2R)X58-12 | +++ | Region 12 |
| 192 | 59A1–3; 59B1–2 | Df(2R)59AB | ++ | Region 12 |
| 193 | 59A1–3; 59D1–4 | Df(2R)59AD | − | |
| 194 | 59D 4–8; 59D9–E1 | Df(2R)vir-12 | − | |
| 195 | 59D 8; 60A7 | Df(2R)bw-S46 | + | FP6 |
| 196 | 59D11; 59F6–8 | Df(2R)bw-HB132 | + | FP4 |
| 197 | 59E; 60A1 | Df(2R)egl2 | ++ | FP1 |
| 198 | 59E1; 59F6 | Df(2R)bw5 | + | FP6 |
| 199 | 59E1; 60C7–D1 | Df(2R)bwVDe2LPxKR | − | |
| 200 | 59F1; 59F5 | Df(2R)egl3 | ++ | FP1 |
| 201 | 59F3; 60A8–16 | Df(2R)G10-7-5 | − | |
| 202 | 59F6; 60A12–16 | Df(2R)or-BR11 | − | |
| 203 | 60B8–10; 60D1 | Df(2R)Px1 | − | |
| 204 | 60C6; 60D9–10 | Df(2R)Px2 | − | |
| 205 | 60E6; 60F1–2 | Df(2R)ES1 | ++ | FP2, -4 |
| 206 | 60E1–2; 60E6 | Df(2R)Dll-MP | ++ | FP4 |
| 207 | 60E10; 60F5 | Df(2R)Kr10 | ++ | FP4 |
| 208 | 60E6–9; 60E11 | Df(2R)M60E | − | |
| 209 | 60E9; 60F1 | Df(2R)gsb | + | FP3, -6 |
| 210 | 60F2; 60F5 | Df(2R)Kr14 | − | |
| 211 | 61A; 61D3 | Df(3L)emc-E12 | − | |
| 212 | 61A1; 61B | Df(3L)B71 | − | |
| 213 | 61C1–4; 61F3 | Df(3L)Ar12-1 | + | FP6 |
| 214 | 61C3–4; 61E | Df(3L)Ar11 | + | FP6 |
| 215 | 61C4; 62A8 | Df(3L)Ar14-8 | + | FP6 |
| 216 | 61D3–E1; 61F5–8 | Df(3L)bab-PG | − | |
| 217 | 61F8; 62A3–5 | Df(3L)ru-22 | + | FP6 |
| 218 | 62A10–B1; 62C4–D1 | Df(3L)R-G5 | − | |
| 219 | 62A10–B1; 62D2 | Df(3L)Aprt-1 | − | |
| 220 | 62B9; 62E7 | Df(3L)R-G7 | − | |
| 221 | 63C1; 63D3 | Df(3L)HR232 | − | |
| 222 | 63C1–2; 63F1–2 | Df(3L)1227 | ++ | FP1 |
| 223 | 63C6; 63F7 | Df(3L)HR119 | − | |
| 224 | 63E2; 64B17 | Df(3L)GN50 | − | |
| 225 | 63F6–7; 64C13–15 | Df(3L)GN24 | − | |
| 226 | 64; 65B5–C1 | Dr(3L)CH39 | − | |
| 227 | 64B–C; 65B5–C1 | Df(3L)CH18 | − | |
| 228 | 64C; 65C | Df(3L)ZN47 | − | |
| 229 | 64E1–13; 65C1–D6 | Df(3L)v65c | ++ | FP1 |
| 230 | 65A; 65E1 | Df(3L)W5.4 | − | |
| 231 | 65A2; 65E1 | Df(3L)XDl98 | − | |
| 232 | 65D4–5; 65E4–6 | Df(3L)BSC27 | − | |
| 233 | 65E1–12; 66A17 | Df(3L)RM5-2 | − | |
| 234 | 65F3; 66B10 | Df(3L)pbl-X1 | − | |
| 235 | 66A17–20; 66C1–5 | Df(3L)ZP1 | − | |
| 236 | 66B12–C1; 66D2–4 | Df(3L)BSC13 | ++ | Region 13 |
| 237 | 66B8–9; 66C9–10 | Df(3L)66C-G28 | +++ | Region 13 |
| 238 | 66C7–10; 66C7–10 | Df(3L)66C-I65 | − | |
| 239 | 66E1–6; 66F1–6 | Df(3L)Scf-R6 | − | |
| 240 | 66E3–4; 66F1–2 | Df(3L)Scf-R11 | − | |
| 241 | 66F5; 66F5 | Df(3L)Rdl-2 | − | |
| 242 | 66F5; 67B1 | Df(3L)29A6 | ++ | Region 14 |
| 243 | 67A2; 67D13 | Df(3L)AC1 | + | FP6 |
| 244 | 67E1–2; 68C1–2 | Df(3L)lxd6 | − | |
| 245 | 68A2; 69A1 | Df(3L)vin5 | − | |
| 246 | 68C8; 69B4–5 | Df(3L)vin7 | − | |
| 247 | 69A4–5; 69D4–6 | Df(3L)eyg-C1 | − | |
| 248 | 69B1–5; 69D1–6 | Df(3L)iro-2 | − | |
| 249 | 69D; 69D | Df(3L)8ex25 | + | FP1, -6 |
| 250 | 69D2; 69E3–5 | Df(3L)E44 | − | |
| 251 | 69D4–5; 69F5–7 | Df(3L)BSC10 | − | |
| 252 | 69F3–4; 70C3–4 | Df(3L)C190LUbx42TR | − | |
| 253 | 70C2; 72A1 | Df(3L)D-5rv12 | − | |
| 254 | 70C2–6; 70E1 | Df(3L)fz-CAL | − | |
| 255 | 70D2; 70E8 | Df(3L)fz-D21 | − | |
| 256 | 70D2; 71E4–5 | Df(3L)fz-M21 | − | |
| 257 | 70E; 71F | Df(3L)Brd6 | ++ | FP1 |
| 258 | 71A1–2; 71C1–2 | Df(3L)Brd15 | − | |
| 259 | 71C2–3; 72B1–C1 | Df(3L)XG-5 | − | |
| 260 | 71C3; 71E5 | Df(3L)BK10 | − | |
| 261 | 72A2; 72D10 | Df(3L)th102 | − | |
| 262 | 72A3–4; 72D1–5 | Df(3L)brm11 | − | |
| 263 | 72C1; 73A4 | Df(3L)st-f13 | − | |
| 264 | 72D10–11; 73D1–2 | Df(3L)st-b11 | − | |
| 265 | 73A3; 74F1–4 | Df(3L)81k19 | − | |
| 266 | 74D3–75A1; 75B2–5 | Df(3L)BSC8 | − | |
| 267 | 75B10; 75C5 | Df(3L)W4 | − | |
| 268 | 75C1–2; 75F1 | Df(3L)Cat | − | |
| 269 | 75F10–11; 76A1–5 | Df(3L)fz2 | − | |
| 270 | 76A7–B1; 76B4–5 | Df(3L)BSC20 | − | |
| 271 | 76B; 76F | Df(3L)XS2182 | − | |
| 272 | 76B; 77A | Df(3L)XS543 | − | |
| 273 | 76B1–2; 76D5 | Df(3L)kto2 | ++ | FP1 |
| 274 | 76B4; 76D3 | Df(3L)XS705 | − | |
| 275 | 76B4; 77B | Df(3L)XS-533 | − | |
| 276 | 76B6; 77C1 | Df(3L)XS572 | − | |
| 277 | 77A1; 77D1 | Df(3L)rdgC-co2 | − | |
| 278 | 77B7–9; 77F1–5 | Df(3L)ri-79c | ++ | Region 15 |
| 279 | 77E2; 78A4 | Df(3L)ri-XT1 | − | |
| 280 | 77F3; 78C8–9 | Df(3L)ME107 | − | |
| 281 | 78C5–6; 78E3–79A1 | Df(3L)Pc-2q | − | |
| 282 | 79C; 79E1–8 | Df(3L)Ten-m-AL29 | − | |
| 283 | 79D3–E1; 79F3–6 | Df(3L)HD1 | − | |
| 284 | 79E1–2; 79E1–8 | Df(3L)Ten-m-AL1 | − | |
| 285 | 79E5–F1; 80A2–3 | Df(3L)BSC21 | − | |
| 286 | 79F; 80A | Df(3L)Delta1AK | − | |
| 287 | 80Fb–g | Df(3L)3-52 | + | FP6 |
| 288 | 80Fd–e | Df(3R)6-61 | + | FP6 |
| 289 | 80Ff–g | Df(3L)8A-80 | − | |
| 290 | 80Fg–j | Df(3L)1-166 | − | |
| 291 | 80Fh–j | Df(3L)2-66 | + | FP6 |
| 292 | 80Fj | Df(3L)2-30 | + | FP6 |
| 293 | 81 Fab | Df(3R)4-75 | − | |
| 294 | 81F; 82F10–11 | Df(3R)2-2 | − | |
| 295 | 81F3–6; 82F5–7 | Df(3R)ME15 | + | FP6 |
| 296 | 82A5–6; 82E4 | Df(3R)Z1 | − | |
| 297 | 82C4; 82F3 | Df(3R)110 | − | |
| 298 | 82D5; 82F3–6 | Df(3R)6-7 | + | FP1, -6 |
| 299 | 82F3–4; 82F10–11 | Df(3R)3-4 | − | |
| 300 | 82F8–10; 83A1–3 | Df(3R)e1025-14 | − | |
| 301 | 83B7–C1; 83C6–D1 | Df(3R)BSC47 | − | |
| 302 | 83E1–2; 84B1 | Df(3R)WIN11 | ++ | Region 16 |
| 303 | 83E3; 84B1 | Df(3R)Dfd13 | +++ | Region 16 |
| 304 | 84A1; 84B1 | Df(3R)9A99 | − | |
| 305 | 84A1–2; 84B1–2 | Df(3R)Scr | − | |
| 306 | 84A6–B1; 84D4–D9 | Df(3R)roe | − | |
| 307 | 84C1–2; 84E1 | Df(3R)dsx2M | − | |
| 308 | 84C8; 84F6 | Df(3R)dsx29 | − | |
| 309 | 84D 4–6; 85B6 | Df(3R)p712 | ++ | FP1 |
| 310 | 84D 8; 85B3–5 | Df(3R)dsx37 | − | |
| 311 | 84D 8–9; 85A1–2 | Df(3R)dsx11 | − | |
| 312 | 84D11; 84E8 | Df(3R)dsx15 | − | |
| 313 | 84E8–9; 85B6 | Df(3R)p40 | ++ | FP1 |
| 314 | 84F1; 85A6–B9 | Df(3R)p13 | − | |
| 315 | 84F2; 85A5–7 | Df(3R)CA3 | ++ | FP1 |
| 316 | 85A2; 85C1–2 | Df(3R)p-XT103 | − | |
| 317 | 85D8; 85E10–13 | Df(3R)by10 | − | |
| 318 | 85D11–13; 85F6 | Df(3R)by62 | +++ | Region 17 |
| 319 | 85D12; 85E10 | Df(3R)GB104 | − | |
| 320 | 86C1; 87B5 | Df(3R)M-Kx1 | − | |
| 321 | 86E2–3; 87C6–7 | Df(3R)T-32 | − | |
| 322 | 86F1–2; 87C6–7 | Df(3R)T-10 | ++ | FP1 |
| 323 | 87B12; 87E8 | Df(3R)ry615 | − | |
| 324 | 87D2; 87F2 | Df(3R)ry27 | − | |
| 325 | 87E–F; 88B | Df(3R)CbxTwtLUbxKM5R | + | FP1, -6 |
| 326 | 87E1; 87F11 | Df(3R)I26c | − | |
| 327 | 87F1; 87F15 | Df(3R)urd | − | |
| 328 | 87F12–14; 88C2 | Df(3R)red31 | − | |
| 329 | 88A2; 88C1–D1 | Df(3R)red1 | − | |
| 330 | 88B1; 88C2 | Df(3R)red-P93 | − | |
| 331 | 88E7–13; 89A1 | Df(3R)ea | + | FP6 |
| 332 | 88F7; 89A11–13 | Df(3R)Po4 | − | |
| 333 | 89A1–2; 89A11–13 | Df(3R)Po2 | ++ | FP1 |
| 334 | 89A1–2; 89A11–13 | Df(3R)Po3 | − | |
| 335 | 89A8; 89B3 | Df(3R)Exel7327 | − | |
| 336 | 89B4; 89B10 | Df(3R)sbd45 | − | |
| 337 | 89B5; 89C | Dr(3R)sd104 | − | |
| 338 | 89B5–6; 89E2–3 | Df(3R)bxd100 | + | FP1, -6 |
| 339 | 89B7–8; 89E7–8 | Df(3R)P115 | − | |
| 340 | 89E1–89F4; 91B1–B2 | Df(3R)DG2 | − | |
| 341 | 89E2–3; 90A | Df(3R)C4 | ++ | FP1 |
| 342 | 89E2–3; 90D | Df(3R)RD31 | − | |
| 343 | 90F1–2; 91F5 | Df(3R)Cha7 | − | |
| 344 | 91A2–B3; 91F13–92A1 | Df(3R)Cha1a | − | |
| 345 | 91F1–2; 92D3–6 | Df(3R)D1-BX12 | − | |
| 346 | 92B3; 92F13 | Df(3R)H-B79 | − | |
| 347 | 93B2–13; 94A3–8 | Df(3R)e-N19 | − | |
| 348 | 93B6; 93D3–4 | Df(3R)e-R1 | − | |
| 349 | 93C6; 94A1–4 | Df(3R)e-GC3 | + | FP1, -6 |
| 350 | 93E–F; 94C–D | Df(3R)5C1 | + | FP1, -6 |
| 351 | 93F11–14; 94D10–13 | Df(3R)hh | − | |
| 352 | 95A5–7; 95C10–11 | Df(3R)mbc-30 | ++ | Region 18 |
| 353 | 95A5–7; 95D6–11 | Df(3R)mbc-R1 | − | |
| 354 | 95D7–11; 95F15 | Df(3R)crb-F89-4 | − | |
| 355 | 95D11–E2; 96A2 | Df(3R)crb87-4 | − | |
| 356 | 96A1; 96A20–25 | Df(3R)Ubx7LLatsR | − | |
| 357 | 96A2–7; 96D2–4 | Df(3R)slo8 | − | |
| 358 | 96F1; 97B1 | Df(3R)Espl3 | − | |
| 359 | 96F10–11; 96F11 | Df(3R)Espl22 | − | |
| 360 | 96F12–14; 97C4–5 | Df(3R)ME61 | − | |
| 361 | 97A; 98A1–2 | Df(3R)T1-P | − | |
| 362 | 97E3; 98A5 | Df(3R)D605 | − | |
| 363 | 98D3–7; 98D3–7 | Df(3R)M15 | − | |
| 364 | 98E3; 99A6 | Df(3R)3450 | − | |
| 365 | 98F14; 99E2–3 | Df(3R)R133 | − | |
| 366 | 99A6; 99C1 | Df(3R)01215 | − | |
| 367 | 99D1–2; 99E1 | Df(3R)X3F | − | |
| 368 | 99F1–2; 100B5 | Df(3R)tll-g | − | |
| 369 | 100A2; 100C2–3 | Df(3R)tll-e | + | FP6 |
| 370 | 100D1; 100D3–4 | Df(3R)awd-KRB | − | |
| 371 | 100D1–2; 100E–F | Df(3R)faf-BP | − | |
| 372 | 100D2; 100F5 | Df(3R)04661 | − |
Breakpoints are as determined by FlyBaseConsortium (2003); otherwise breakpoints were supplied by the Bloomington stock center.
−, nonsuppressor; +, equivocal suppressor with a phenotype that overlaps the nonsuppressed phenotype; ++, weak suppressor; +++, moderate suppressor; ++++, strong suppressor (see Figure 1).
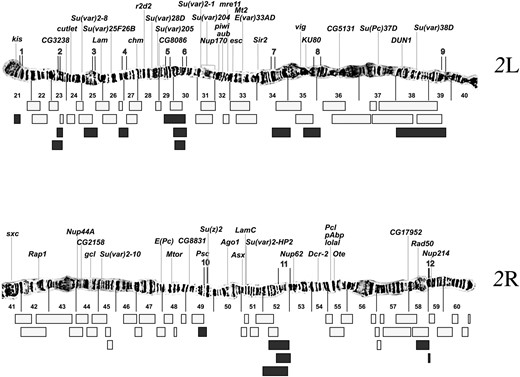
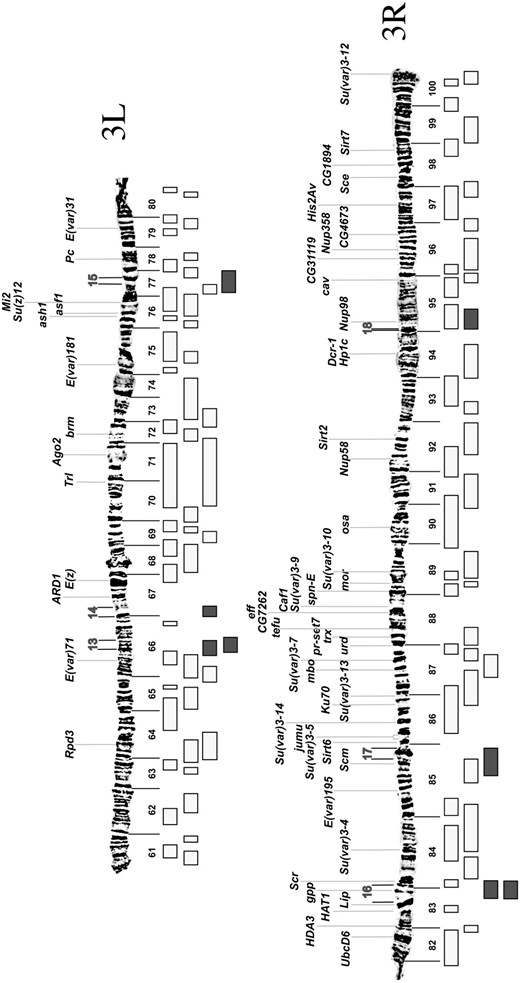
Determination of suppressor on the deficiency chromosome. FP1, false positive because the region of the deficiency is covered by one or more nonsuppressing deficiencies; FP2, false positive because the suppressor maps to the tip of 2L rather than to the site of the deficiency; FP3, false positive because the region of the deficiency is covered by a combination of nonsuppressing deficiencies and deficiencies with suppressors that map to the 2L tip; FP4, assumed false positive because the 2L TAS is missing by in situ hybridization; FP5, assumed false positive because the deficiency chromosome fails to complement lethal mutations of l(2)gl; FP6, the suppressor cannot be adequately tested because the phenotype overlaps wild type. Regions refer to map positions as shown in Figures 3 and 4.
Deficiency chromosomes tested for suppression of telomeric silencing
Df no. . | Breakpointsa . | Name . | Suppressionb . | Callc . |
|---|---|---|---|---|
| 1 | 21A1; 21B6–7 | Df(2L)net-PM47C | ++ | FP4 |
| 2 | 21A1; 21B7–8 | Df(2L)net-PMF | + | FP4, -5 |
| 3 | 21A1–4; 21B3 | Df(2L)net18 | ++ | FP4, -5 |
| 4 | 21A1–4; 21B4 | Df(2L)net62 | ++ | FP4, -5 |
| 5 | 21C1; 21C7 | Df(2L)al | ++ | Region 1 |
| 6 | 21C3–4; 21C6–8 | Df(2L)BSC16 | − | |
| 7 | 21C8–D1; 22A8–B1 | Df(2L)S2 | +++ | FP4 |
| 8 | 21D1–2; 21E1–2 | Df(2L)ast4 | +++ | FP4, -5 |
| 9 | 21D1–2; 22B2–3 | Df(2L)ast2 | + | FP6 |
| 10 | 21D2; 21F3–22A1 | Df(2L)S3 | + | FP6 |
| 11 | 21E3–4; 22B5–7 | Df(2L)frtz17 | − | |
| 12 | 22A1; 22B6–9 | D(2L)frtz11 | ++ | FP1 |
| 13 | 22A2–3; 22B7 | D(2L)frtz19 | ++ | FP1 |
| 14 | 22A2–3; 22D5–E1 | Df(2L)dp-79b | − | |
| 15 | 22A3; 22B3 | Df(2L)frtz14 | + | FP1, -6 |
| 16 | 22A3–4; 22C1–2 | Df(2L)frtz25 | +++ | FP1 |
| 17 | 22A6; 22B9 | Df(2L)J69LH56R | ++ | FP1 |
| 18 | 22F1–2; 23A2 | Df(2L)dpp-d14 | − | |
| 19 | 22F3–4; 23C3–5 | Df(2L)C144 | − | |
| 20 | 22F4; 23A1 | Df(2L)D20 | ++ | FP1, -4 |
| 21 | 23A3–4; 23D4–6 | Df(2L)JS13 | ++ | Region 2 |
| 22 | 23C1–2; 23E1–2 | Df(2L)JS17 | ++ | Region 2 |
| 23 | 23C3–5; 23D1–2 | Df(2L)JS32 | − | |
| 24 | 23D2; 23E3 | Df(2L)S2590 | ++ | FP2, -4 |
| 25 | 23F2–3; 23F6–24A1 | Df(2L)tim-02 | − | |
| 26 | 24A2; 24D4 | Df(2L)ed1 | − | |
| 27 | 24C3; 25A2 | Df(2L)ed-dp | + | FP6 |
| 28 | 24E1; 25A2 | Df(2L)M24F-B | − | |
| 29 | 24E3; 25A7 | Df(2L)sc19-3 | − | |
| 30 | 24E4; 25B2 | Df(2L)dp-h25 | − | |
| 31 | 24F1–2; 25C5 | Df(2L)sc19-6 | − | |
| 32 | 25A5; 25D6 | Df(2L)sc19-5 | ++ | Region 3 |
| 33 | 25D2–3; 26B2–5 | Df(2L)cl-h3 | − | |
| 34 | 25F3–26A1; 26D3–11 | Df(2L)E110 | − | |
| 35 | 26D3–E1; 26F4–7 | Df(2L)BSC6 | − | |
| 36 | 26D10–E1; 27C1 | Df(2L)BSC7 | ++ | Region 4 |
| 37 | 27B2; 27F1–2 | Df(2L)spd-j2 | − | |
| 38 | 27C5–9; 28B3–4 | Df(2L)J-H | − | |
| 39 | 27E; 28C1–4 | Df(2L)spd | + | FP6 |
| 40 | 27E2; 28D1 | Df(2L)XE-3801 | +++ | FP4 |
| 41 | 28B2; 28D1 | Df(2L)XE-2750 | + | FP6 |
| 42 | 28DE (within) | Df(2L)Trf-C6R31 | ++ | FP4 |
| 43 | 28E4–7; 29B2–C1 | Df(2L)TE29Aa-11 | − | |
| 44 | 29C1–2; 30C8–9 | Df(2L)N22-14 | ++ | FP4 |
| 45 | 29C3–5; 30C8–9 | Df(2L)N22-5 | ++ | Regions 5, 6 |
| 46 | 29E2–F1; 30C2–4 | Df(2L)TE30Cb-1 | − | |
| 47 | 30A1–2; 30D1–2 | Df(2L)N22-3 | ++ | Region 6 |
| 48 | 30A3–5; 30C5 | Df(2L)30A-C | ++ | Region 6 |
| 49 | 30A9–B1; 30D2–F4 | Df(2L)gamma7 | +++ | FP4 |
| 50 | 30D1–F6; 31F1–5 | Df(2L)Mdh | − | |
| 51 | 31B1; 32A1–2 | Df(2L)J2 | − | |
| 52 | 31D1–11; 31E7 | Df(2L)J27 | − | |
| 53 | 32D1; 32F1–3 | Df(2L)FCK-20 | − | |
| 54 | 32F1–3; 33F1–2 | Df(2L)Prl | + | FP5, -6 |
| 55 | 33A1; 33B1–2 | Df(2L)esc-P2-0 | + | FP6 |
| 56 | 33A1; 33B2 | Df(2L)esc10 | − | |
| 57 | 33A1; 33E | Df(2L)esc-P3-0 | − | |
| 58 | 33B3; 34A1–2 | Df(2L)prd1.7 | − | |
| 59 | 34B7–12; 34E3 | In(2L)b82a1 | ++ | Region 7 |
| 60 | 34C1; 35C1 | Df(2L)b87e25 | ++ | FP2, -5 |
| 61 | 34C4; 35A4 | Df(2L)b80e3 | − | |
| 62 | 34D1–2; 35C1 | Df(2L)64j | +++ | FP1 |
| 63 | 34D3–4; 35C1 | Df(2L)fn30 | ++++ | FP1 |
| 64 | 34D4; 34E3 | Df(2L)b88f32 | ++ | FP1 |
| 65 | 34E2; 35B3–4 | Df(2L)fn7 | +++ | FP1 |
| 66 | 34E3; 35D2–5 | Df(2L)el80f1 | + | FP1, -6 |
| 67 | 34E4–34F1; 35C3 | Df(2L)noc11 | − | |
| 68 | 34E5–F1; 35C3 | Df(2L)A263 | − | |
| 69 | 34F2–5; 35C4 | Df(2L)osp38 | ++ | FP1 |
| 70 | 34F4; 35C3 | Df(2L)fn5 | − | |
| 71 | 34F4–5; 35D4–5 | Df(2L)fn1 | ++++ | FP1 |
| 72 | 34F5; 35B2 | Df(2L)el81i1 | − | |
| 73 | 34F5; 35B10 | Df(2L)TE35BC-31 | − | |
| 74 | 34F5–35A4; 35D2 | Df(2L)do1 | − | |
| 75 | 35A1–4; 35C1–3 | Df(2L)A400 | − | |
| 76 | 35A4–B1; 35B2 | Df(2L)fn2 | + | FP1 |
| 77 | 35B1; 35F1 | Df(2L)A446 | − | |
| 78 | 35B3; 35E6 | Df(2L)osp29 | − | |
| 79 | 35B4–6; 35E1–2 | Df(2L)TE35BC-24 | − | |
| 80 | 35D1; 36A6–7 | Df(2L)r10 | ++ | Region 8 |
| 81 | 35F6–12; 36D | Df(2L)cact-255rv64 | +++ | FP5 |
| 82 | 36A8–9; 36F1 | Df(2L)H20 | − | |
| 83 | 36C2–4; 37B9–10 | Df(2L)TW137 | − | |
| 84 | 36E4–F1; 38A6–7 | Df(2L)TW50 | +++ | FP1 |
| 85 | 36F7–9; 37B2–7 | Df(2L)TW3 | − | |
| 86 | 36F7–9; 37D1–2 | Df(2L)VA16 | ++ | FP1 |
| 87 | 37B2–8; 37C5 | Df(2L)hk-UC2 | − | |
| 88 | 37B2–10; 38D2–5 | Df(2L)pr-A16 | − | |
| 89 | 37B2–8; 37E2 | Df(2L)TW158 | ++ | FP1 |
| 90 | 37B9–10; 37D1–2 | Df(2L)TW130 | ++ | FP1 |
| 91 | 37B9–10; 37D5 | Df(2L)VA23 | ++ | FP1 |
| 92 | 37C1; 37F5 | Df(2L)VA17 | ++ | FP1 |
| 93 | 37C2–5; 38B2–C1 | Df(2L)VA12 | +++ | FP1 |
| 94 | 37C2–7; 38C1–2 | Df(2L)Sd77 | − | |
| 95 | 37D2; 38A1 | Df(2L)E55 | +++ | FP1 |
| 96 | 37D2–5; 38A6–B2 | Df(2L)Sd37 | − | |
| 97 | 37D2–5; 39A4–7 | Df(2L)pr-A14 | ++ | FP1 |
| 98 | 37D6–E1; 38E6–9 | Df(2L)TW2 | ++ | FP1 |
| 99 | 37E2–4; 39D1 | Df(2L)TW12 | − | |
| 100 | 37E2–F1; 38B5–C1 | Df(2L)TW9 | +++ | FP1 |
| 101 | 38A1; 39D3–E1 | Df(2L)TW84 | + | FP1, -6 |
| 102 | 38A1; 39F1 | Df(2L)TW65 | ++ | Region 9 |
| 103 | 38A3–4; 38B6–C1 | Df(2L)pr-A20 | ++ | FP1 |
| 104 | 38A7–B1; 39C2–3 | Df(2L)TW1 | ++ | FP1 |
| 105 | 38B3–6; 40A3 | Df(2L)pr-M1 | + | FP6 |
| 106 | 38E2; 39E7 | Df(2L)DS6 | − | |
| 107 | 40h35; 40h38L | Df(2L)C' | − | |
| 108 | h38R; h46 | Df(2R)M41A10 | + | FP6 |
| 109 | h42–h43; 42A2–3 | In(2R)bwVDe2LCyR | + | FP6 |
| 110 | h44–h46; 41B1–41F11 | Df(2R)M41A8 | +++ | FP4, -5 |
| 111 | h44–h46; 42A1–2 | Df(2R)M41A4 | ++ | FP4 |
| 112 | 41BC; 42A16–B1 | Df(2R)nap14 | + | FP6 |
| 113 | 41D2–E1; 42B1–3 | Df(2R)nap1 | − | |
| 114 | 41F3–4; 42A3–9 | Df(2R)17I | ++ | FP1 |
| 115 | 42A1–2; 42E6–F1 | Df(2R)nap9 | − | |
| 116 | 42A1–19; 42E2–7 | Df(2R)cn88b | − | |
| 117 | 42B3–4; 43E18 | Df(2R)ST1 | ++ | FP1 |
| 118 | 42B4–C1; 43F–44A1 | Df(2R)cn87e | − | |
| 119 | 42C1–7; 43F5–8 | Df(2R)pk78s | − | |
| 120 | 42C2; 42D2–3 | Df(2R)42 | − | |
| 121 | 42C2–7; 43D1–7 | Df(2R)Drl-rv17 | + | FP1, -6 |
| 122 | 42E; 44C1 | Df(2R)cn9 | ++ | FP1 |
| 123 | 42E1–4; 43C3 | Df(2R)Drl-rv3 | ++ | FP1 |
| 124 | 43A3; 43F6 | Df(2R)P32a | + | FP1, -6 |
| 125 | 43C5; 44B6–C1 | Df(2R)cn83c | + | FP1, -6 |
| 126 | 43C7; 43F2–8 | Df(2R)cn-S6 | − | |
| 127 | 43E7–18; 44B4–5 | Df(2R)CA53 | − | |
| 128 | 43F; 44D3–8 | Df(2R)H3C1 | − | |
| 129 | 44C1–2; 44E1–4 | Df(2R)44CE | − | |
| 130 | 44D1–4; 44F12 | Df(2R)H3E1 | − | |
| 131 | 44D2–E1; 45B8–C1 | Df(2R)Np3 | − | |
| 132 | 44F11; 45C1 | Df(2R)Np4 | ++ | FP1 |
| 133 | 44F11; 45D9–E1 | Df(2R)Np5 | + | FP1, -6 |
| 134 | 44F2–3; 45C6 | Df(2R)Np1 | − | |
| 135 | 45A6–7; 45E2–3 | Df(2R)w45-30n | + | FP4 |
| 136 | 45A9–10; 45D5–8 | Df(2R)w73-1 | − | |
| 137 | 45C8; 45D8 | Df(2R)wun-GL | ++ | FP1 |
| 138 | 45C8–D10; 45D9–E1 | Df(2R)w45-19g | ++ | FP1 |
| 139 | 45D3–4; 45F2–6 | Df(2R)BSC29 | − | |
| 140 | 46A1–4; 46C3–12 | Df(2R)B5 | + | FP6 |
| 141 | 46C1–2; 46E1–2 | Df(2R)X3 | − | |
| 142 | 46C2; 47A1 | Df(2R)X1 | − | |
| 143 | 46C3–4; 46C9–11 | Df(2R)eve | +++ | FP1, -2 |
| 144 | 46F1; 47A10 | Df(2R)12 | ++ | FP2 |
| 145 | 46F1; 47B9 | Df(2R)stan2 | ++ | FP3 |
| 146 | 47A3; 47E | Df(2R)E3363 | − | |
| 147 | 47D3; 48A5 | Df(2R)en-A | + | FP6 |
| 148 | 47E3; 48A5–B2 | Df(2R)en-B | ++ | FP2, -4 |
| 149 | 48A1; 48B5 | Df(2R)en-SFX31 | +++ | FP3 |
| 150 | 48A1–2; 48B–C1 | Df(2R)en28 | + | FP3, -6 |
| 151 | 48A3; 48C6–8 | Df(2R)en30 | − | |
| 152 | 48C5–D1; 48D5–E11 | Df(2R)BSC39 | − | |
| 153 | 48E; 49A | Df(2R)CB21 | +++ | FP2, -5 |
| 154 | 49A; 49E1–2 | Df(2R)vg135 | + | FP5, -6 |
| 155 | 49B2–3; 49E2 | Df(2R)vg-C | − | |
| 156 | 49C1–2; 49E6 | Df(2R)vg-D | ++ | Region 10 |
| 157 | 49C1–4; 50C23–D1 | Df(2R)CX1 | +++ | FP4 |
| 158 | 49D3–4; 50A2 | Df(2R)vg-B | + | FP6 |
| 159 | 50C21–23; 50D1–5 | Df(2R)50C-101 | − | |
| 160 | 50E6–F1; 51E2–4 | Df(2R)BSC11 | +++ | FP4 |
| 161 | 50F6–9; 51B3 | Df(2R)L48 | − | |
| 162 | 51A2; 51B6 | Df(2R)trix | ++ | FP1 |
| 163 | 51A5; 51C1 | Df(2R)03072 | − | |
| 164 | 51C3–7; 51E7–11 | Df(2R)14 | − | |
| 165 | 51D1–2; 51E5 | Df(2R)XTE-58 | − | |
| 166 | 51D3–E1; 52D1 | Df(2R)XTE-18 | − | |
| 167 | 51F13; 52F8–9 | Df(2R)Jp4 | +++ | Region 11 |
| 168 | 52A13–14; 52F10–11 | Df(2R)Jp5 | +++ | Region 11 |
| 169 | 52A9–10; 52D9–15 | Df(2R)WMG | − | |
| 170 | 52D3; 53A1 | Df(2R)Jp6 | ++ | Region 11 |
| 171 | 54E8–F1; 55B9–C1 | Df(2R)Pcl7B | − | |
| 172 | 54F2; 56A1 | Df(2R)RM2-1 | +++ | FP3 |
| 173 | 55A1; 55C1–3 | Df(2R)Pcl11B | − | |
| 174 | 55A1; 55F1–2 | Df(2R)PC4 | − | |
| 175 | 55C1–2; 56B1–2 | Df(2R)C29 | ++ | FP3 |
| 176 | 55D2–E1; 56B2 | Df(2R)PC66 | ++ | FP3 |
| 177 | 55E6–F3; 56C1 | Df(2R)P34 | ++ | FP2 |
| 178 | 56D7–E3; 56F9–12 | Df(2R)BSC22 | + | FP6 |
| 179 | 56F 5; 56F15 | Df(2R)173 | − | |
| 180 | 56F 5; 56F15 | Df(2R)017 | − | |
| 181 | 56F 9–11; 57D12 | Df(2R)AA21 | + | FP3, -6 |
| 182 | 56F12–14; 57A4 | Df(2R)BSC19 | − | |
| 183 | 57A3; 57B1 | Df(2R)exu2 | +++ | FP2 |
| 184 | 57A6; 57B6 | Df(2R)D4 | − | |
| 185 | 57B1; 57B13–14 | Df(2R)E2 | − | |
| 186 | 57B4; 58B1–2 | Df(2R)Pu-D17 | − | |
| 187 | 57D2–8; 58D1 | Df(2R)Egfr5 | − | |
| 188 | 58B1–2; 58E4 | Df(2R)X58-7 | − | |
| 189 | 58B3; 59A1 | Df(2R)X58-8 | − | |
| 190 | 58C3–7; 58D6–8 | Df(2R)X58-3 | − | |
| 191 | 58D1–2; 59A1 | Df(2R)X58-12 | +++ | Region 12 |
| 192 | 59A1–3; 59B1–2 | Df(2R)59AB | ++ | Region 12 |
| 193 | 59A1–3; 59D1–4 | Df(2R)59AD | − | |
| 194 | 59D 4–8; 59D9–E1 | Df(2R)vir-12 | − | |
| 195 | 59D 8; 60A7 | Df(2R)bw-S46 | + | FP6 |
| 196 | 59D11; 59F6–8 | Df(2R)bw-HB132 | + | FP4 |
| 197 | 59E; 60A1 | Df(2R)egl2 | ++ | FP1 |
| 198 | 59E1; 59F6 | Df(2R)bw5 | + | FP6 |
| 199 | 59E1; 60C7–D1 | Df(2R)bwVDe2LPxKR | − | |
| 200 | 59F1; 59F5 | Df(2R)egl3 | ++ | FP1 |
| 201 | 59F3; 60A8–16 | Df(2R)G10-7-5 | − | |
| 202 | 59F6; 60A12–16 | Df(2R)or-BR11 | − | |
| 203 | 60B8–10; 60D1 | Df(2R)Px1 | − | |
| 204 | 60C6; 60D9–10 | Df(2R)Px2 | − | |
| 205 | 60E6; 60F1–2 | Df(2R)ES1 | ++ | FP2, -4 |
| 206 | 60E1–2; 60E6 | Df(2R)Dll-MP | ++ | FP4 |
| 207 | 60E10; 60F5 | Df(2R)Kr10 | ++ | FP4 |
| 208 | 60E6–9; 60E11 | Df(2R)M60E | − | |
| 209 | 60E9; 60F1 | Df(2R)gsb | + | FP3, -6 |
| 210 | 60F2; 60F5 | Df(2R)Kr14 | − | |
| 211 | 61A; 61D3 | Df(3L)emc-E12 | − | |
| 212 | 61A1; 61B | Df(3L)B71 | − | |
| 213 | 61C1–4; 61F3 | Df(3L)Ar12-1 | + | FP6 |
| 214 | 61C3–4; 61E | Df(3L)Ar11 | + | FP6 |
| 215 | 61C4; 62A8 | Df(3L)Ar14-8 | + | FP6 |
| 216 | 61D3–E1; 61F5–8 | Df(3L)bab-PG | − | |
| 217 | 61F8; 62A3–5 | Df(3L)ru-22 | + | FP6 |
| 218 | 62A10–B1; 62C4–D1 | Df(3L)R-G5 | − | |
| 219 | 62A10–B1; 62D2 | Df(3L)Aprt-1 | − | |
| 220 | 62B9; 62E7 | Df(3L)R-G7 | − | |
| 221 | 63C1; 63D3 | Df(3L)HR232 | − | |
| 222 | 63C1–2; 63F1–2 | Df(3L)1227 | ++ | FP1 |
| 223 | 63C6; 63F7 | Df(3L)HR119 | − | |
| 224 | 63E2; 64B17 | Df(3L)GN50 | − | |
| 225 | 63F6–7; 64C13–15 | Df(3L)GN24 | − | |
| 226 | 64; 65B5–C1 | Dr(3L)CH39 | − | |
| 227 | 64B–C; 65B5–C1 | Df(3L)CH18 | − | |
| 228 | 64C; 65C | Df(3L)ZN47 | − | |
| 229 | 64E1–13; 65C1–D6 | Df(3L)v65c | ++ | FP1 |
| 230 | 65A; 65E1 | Df(3L)W5.4 | − | |
| 231 | 65A2; 65E1 | Df(3L)XDl98 | − | |
| 232 | 65D4–5; 65E4–6 | Df(3L)BSC27 | − | |
| 233 | 65E1–12; 66A17 | Df(3L)RM5-2 | − | |
| 234 | 65F3; 66B10 | Df(3L)pbl-X1 | − | |
| 235 | 66A17–20; 66C1–5 | Df(3L)ZP1 | − | |
| 236 | 66B12–C1; 66D2–4 | Df(3L)BSC13 | ++ | Region 13 |
| 237 | 66B8–9; 66C9–10 | Df(3L)66C-G28 | +++ | Region 13 |
| 238 | 66C7–10; 66C7–10 | Df(3L)66C-I65 | − | |
| 239 | 66E1–6; 66F1–6 | Df(3L)Scf-R6 | − | |
| 240 | 66E3–4; 66F1–2 | Df(3L)Scf-R11 | − | |
| 241 | 66F5; 66F5 | Df(3L)Rdl-2 | − | |
| 242 | 66F5; 67B1 | Df(3L)29A6 | ++ | Region 14 |
| 243 | 67A2; 67D13 | Df(3L)AC1 | + | FP6 |
| 244 | 67E1–2; 68C1–2 | Df(3L)lxd6 | − | |
| 245 | 68A2; 69A1 | Df(3L)vin5 | − | |
| 246 | 68C8; 69B4–5 | Df(3L)vin7 | − | |
| 247 | 69A4–5; 69D4–6 | Df(3L)eyg-C1 | − | |
| 248 | 69B1–5; 69D1–6 | Df(3L)iro-2 | − | |
| 249 | 69D; 69D | Df(3L)8ex25 | + | FP1, -6 |
| 250 | 69D2; 69E3–5 | Df(3L)E44 | − | |
| 251 | 69D4–5; 69F5–7 | Df(3L)BSC10 | − | |
| 252 | 69F3–4; 70C3–4 | Df(3L)C190LUbx42TR | − | |
| 253 | 70C2; 72A1 | Df(3L)D-5rv12 | − | |
| 254 | 70C2–6; 70E1 | Df(3L)fz-CAL | − | |
| 255 | 70D2; 70E8 | Df(3L)fz-D21 | − | |
| 256 | 70D2; 71E4–5 | Df(3L)fz-M21 | − | |
| 257 | 70E; 71F | Df(3L)Brd6 | ++ | FP1 |
| 258 | 71A1–2; 71C1–2 | Df(3L)Brd15 | − | |
| 259 | 71C2–3; 72B1–C1 | Df(3L)XG-5 | − | |
| 260 | 71C3; 71E5 | Df(3L)BK10 | − | |
| 261 | 72A2; 72D10 | Df(3L)th102 | − | |
| 262 | 72A3–4; 72D1–5 | Df(3L)brm11 | − | |
| 263 | 72C1; 73A4 | Df(3L)st-f13 | − | |
| 264 | 72D10–11; 73D1–2 | Df(3L)st-b11 | − | |
| 265 | 73A3; 74F1–4 | Df(3L)81k19 | − | |
| 266 | 74D3–75A1; 75B2–5 | Df(3L)BSC8 | − | |
| 267 | 75B10; 75C5 | Df(3L)W4 | − | |
| 268 | 75C1–2; 75F1 | Df(3L)Cat | − | |
| 269 | 75F10–11; 76A1–5 | Df(3L)fz2 | − | |
| 270 | 76A7–B1; 76B4–5 | Df(3L)BSC20 | − | |
| 271 | 76B; 76F | Df(3L)XS2182 | − | |
| 272 | 76B; 77A | Df(3L)XS543 | − | |
| 273 | 76B1–2; 76D5 | Df(3L)kto2 | ++ | FP1 |
| 274 | 76B4; 76D3 | Df(3L)XS705 | − | |
| 275 | 76B4; 77B | Df(3L)XS-533 | − | |
| 276 | 76B6; 77C1 | Df(3L)XS572 | − | |
| 277 | 77A1; 77D1 | Df(3L)rdgC-co2 | − | |
| 278 | 77B7–9; 77F1–5 | Df(3L)ri-79c | ++ | Region 15 |
| 279 | 77E2; 78A4 | Df(3L)ri-XT1 | − | |
| 280 | 77F3; 78C8–9 | Df(3L)ME107 | − | |
| 281 | 78C5–6; 78E3–79A1 | Df(3L)Pc-2q | − | |
| 282 | 79C; 79E1–8 | Df(3L)Ten-m-AL29 | − | |
| 283 | 79D3–E1; 79F3–6 | Df(3L)HD1 | − | |
| 284 | 79E1–2; 79E1–8 | Df(3L)Ten-m-AL1 | − | |
| 285 | 79E5–F1; 80A2–3 | Df(3L)BSC21 | − | |
| 286 | 79F; 80A | Df(3L)Delta1AK | − | |
| 287 | 80Fb–g | Df(3L)3-52 | + | FP6 |
| 288 | 80Fd–e | Df(3R)6-61 | + | FP6 |
| 289 | 80Ff–g | Df(3L)8A-80 | − | |
| 290 | 80Fg–j | Df(3L)1-166 | − | |
| 291 | 80Fh–j | Df(3L)2-66 | + | FP6 |
| 292 | 80Fj | Df(3L)2-30 | + | FP6 |
| 293 | 81 Fab | Df(3R)4-75 | − | |
| 294 | 81F; 82F10–11 | Df(3R)2-2 | − | |
| 295 | 81F3–6; 82F5–7 | Df(3R)ME15 | + | FP6 |
| 296 | 82A5–6; 82E4 | Df(3R)Z1 | − | |
| 297 | 82C4; 82F3 | Df(3R)110 | − | |
| 298 | 82D5; 82F3–6 | Df(3R)6-7 | + | FP1, -6 |
| 299 | 82F3–4; 82F10–11 | Df(3R)3-4 | − | |
| 300 | 82F8–10; 83A1–3 | Df(3R)e1025-14 | − | |
| 301 | 83B7–C1; 83C6–D1 | Df(3R)BSC47 | − | |
| 302 | 83E1–2; 84B1 | Df(3R)WIN11 | ++ | Region 16 |
| 303 | 83E3; 84B1 | Df(3R)Dfd13 | +++ | Region 16 |
| 304 | 84A1; 84B1 | Df(3R)9A99 | − | |
| 305 | 84A1–2; 84B1–2 | Df(3R)Scr | − | |
| 306 | 84A6–B1; 84D4–D9 | Df(3R)roe | − | |
| 307 | 84C1–2; 84E1 | Df(3R)dsx2M | − | |
| 308 | 84C8; 84F6 | Df(3R)dsx29 | − | |
| 309 | 84D 4–6; 85B6 | Df(3R)p712 | ++ | FP1 |
| 310 | 84D 8; 85B3–5 | Df(3R)dsx37 | − | |
| 311 | 84D 8–9; 85A1–2 | Df(3R)dsx11 | − | |
| 312 | 84D11; 84E8 | Df(3R)dsx15 | − | |
| 313 | 84E8–9; 85B6 | Df(3R)p40 | ++ | FP1 |
| 314 | 84F1; 85A6–B9 | Df(3R)p13 | − | |
| 315 | 84F2; 85A5–7 | Df(3R)CA3 | ++ | FP1 |
| 316 | 85A2; 85C1–2 | Df(3R)p-XT103 | − | |
| 317 | 85D8; 85E10–13 | Df(3R)by10 | − | |
| 318 | 85D11–13; 85F6 | Df(3R)by62 | +++ | Region 17 |
| 319 | 85D12; 85E10 | Df(3R)GB104 | − | |
| 320 | 86C1; 87B5 | Df(3R)M-Kx1 | − | |
| 321 | 86E2–3; 87C6–7 | Df(3R)T-32 | − | |
| 322 | 86F1–2; 87C6–7 | Df(3R)T-10 | ++ | FP1 |
| 323 | 87B12; 87E8 | Df(3R)ry615 | − | |
| 324 | 87D2; 87F2 | Df(3R)ry27 | − | |
| 325 | 87E–F; 88B | Df(3R)CbxTwtLUbxKM5R | + | FP1, -6 |
| 326 | 87E1; 87F11 | Df(3R)I26c | − | |
| 327 | 87F1; 87F15 | Df(3R)urd | − | |
| 328 | 87F12–14; 88C2 | Df(3R)red31 | − | |
| 329 | 88A2; 88C1–D1 | Df(3R)red1 | − | |
| 330 | 88B1; 88C2 | Df(3R)red-P93 | − | |
| 331 | 88E7–13; 89A1 | Df(3R)ea | + | FP6 |
| 332 | 88F7; 89A11–13 | Df(3R)Po4 | − | |
| 333 | 89A1–2; 89A11–13 | Df(3R)Po2 | ++ | FP1 |
| 334 | 89A1–2; 89A11–13 | Df(3R)Po3 | − | |
| 335 | 89A8; 89B3 | Df(3R)Exel7327 | − | |
| 336 | 89B4; 89B10 | Df(3R)sbd45 | − | |
| 337 | 89B5; 89C | Dr(3R)sd104 | − | |
| 338 | 89B5–6; 89E2–3 | Df(3R)bxd100 | + | FP1, -6 |
| 339 | 89B7–8; 89E7–8 | Df(3R)P115 | − | |
| 340 | 89E1–89F4; 91B1–B2 | Df(3R)DG2 | − | |
| 341 | 89E2–3; 90A | Df(3R)C4 | ++ | FP1 |
| 342 | 89E2–3; 90D | Df(3R)RD31 | − | |
| 343 | 90F1–2; 91F5 | Df(3R)Cha7 | − | |
| 344 | 91A2–B3; 91F13–92A1 | Df(3R)Cha1a | − | |
| 345 | 91F1–2; 92D3–6 | Df(3R)D1-BX12 | − | |
| 346 | 92B3; 92F13 | Df(3R)H-B79 | − | |
| 347 | 93B2–13; 94A3–8 | Df(3R)e-N19 | − | |
| 348 | 93B6; 93D3–4 | Df(3R)e-R1 | − | |
| 349 | 93C6; 94A1–4 | Df(3R)e-GC3 | + | FP1, -6 |
| 350 | 93E–F; 94C–D | Df(3R)5C1 | + | FP1, -6 |
| 351 | 93F11–14; 94D10–13 | Df(3R)hh | − | |
| 352 | 95A5–7; 95C10–11 | Df(3R)mbc-30 | ++ | Region 18 |
| 353 | 95A5–7; 95D6–11 | Df(3R)mbc-R1 | − | |
| 354 | 95D7–11; 95F15 | Df(3R)crb-F89-4 | − | |
| 355 | 95D11–E2; 96A2 | Df(3R)crb87-4 | − | |
| 356 | 96A1; 96A20–25 | Df(3R)Ubx7LLatsR | − | |
| 357 | 96A2–7; 96D2–4 | Df(3R)slo8 | − | |
| 358 | 96F1; 97B1 | Df(3R)Espl3 | − | |
| 359 | 96F10–11; 96F11 | Df(3R)Espl22 | − | |
| 360 | 96F12–14; 97C4–5 | Df(3R)ME61 | − | |
| 361 | 97A; 98A1–2 | Df(3R)T1-P | − | |
| 362 | 97E3; 98A5 | Df(3R)D605 | − | |
| 363 | 98D3–7; 98D3–7 | Df(3R)M15 | − | |
| 364 | 98E3; 99A6 | Df(3R)3450 | − | |
| 365 | 98F14; 99E2–3 | Df(3R)R133 | − | |
| 366 | 99A6; 99C1 | Df(3R)01215 | − | |
| 367 | 99D1–2; 99E1 | Df(3R)X3F | − | |
| 368 | 99F1–2; 100B5 | Df(3R)tll-g | − | |
| 369 | 100A2; 100C2–3 | Df(3R)tll-e | + | FP6 |
| 370 | 100D1; 100D3–4 | Df(3R)awd-KRB | − | |
| 371 | 100D1–2; 100E–F | Df(3R)faf-BP | − | |
| 372 | 100D2; 100F5 | Df(3R)04661 | − |
Df no. . | Breakpointsa . | Name . | Suppressionb . | Callc . |
|---|---|---|---|---|
| 1 | 21A1; 21B6–7 | Df(2L)net-PM47C | ++ | FP4 |
| 2 | 21A1; 21B7–8 | Df(2L)net-PMF | + | FP4, -5 |
| 3 | 21A1–4; 21B3 | Df(2L)net18 | ++ | FP4, -5 |
| 4 | 21A1–4; 21B4 | Df(2L)net62 | ++ | FP4, -5 |
| 5 | 21C1; 21C7 | Df(2L)al | ++ | Region 1 |
| 6 | 21C3–4; 21C6–8 | Df(2L)BSC16 | − | |
| 7 | 21C8–D1; 22A8–B1 | Df(2L)S2 | +++ | FP4 |
| 8 | 21D1–2; 21E1–2 | Df(2L)ast4 | +++ | FP4, -5 |
| 9 | 21D1–2; 22B2–3 | Df(2L)ast2 | + | FP6 |
| 10 | 21D2; 21F3–22A1 | Df(2L)S3 | + | FP6 |
| 11 | 21E3–4; 22B5–7 | Df(2L)frtz17 | − | |
| 12 | 22A1; 22B6–9 | D(2L)frtz11 | ++ | FP1 |
| 13 | 22A2–3; 22B7 | D(2L)frtz19 | ++ | FP1 |
| 14 | 22A2–3; 22D5–E1 | Df(2L)dp-79b | − | |
| 15 | 22A3; 22B3 | Df(2L)frtz14 | + | FP1, -6 |
| 16 | 22A3–4; 22C1–2 | Df(2L)frtz25 | +++ | FP1 |
| 17 | 22A6; 22B9 | Df(2L)J69LH56R | ++ | FP1 |
| 18 | 22F1–2; 23A2 | Df(2L)dpp-d14 | − | |
| 19 | 22F3–4; 23C3–5 | Df(2L)C144 | − | |
| 20 | 22F4; 23A1 | Df(2L)D20 | ++ | FP1, -4 |
| 21 | 23A3–4; 23D4–6 | Df(2L)JS13 | ++ | Region 2 |
| 22 | 23C1–2; 23E1–2 | Df(2L)JS17 | ++ | Region 2 |
| 23 | 23C3–5; 23D1–2 | Df(2L)JS32 | − | |
| 24 | 23D2; 23E3 | Df(2L)S2590 | ++ | FP2, -4 |
| 25 | 23F2–3; 23F6–24A1 | Df(2L)tim-02 | − | |
| 26 | 24A2; 24D4 | Df(2L)ed1 | − | |
| 27 | 24C3; 25A2 | Df(2L)ed-dp | + | FP6 |
| 28 | 24E1; 25A2 | Df(2L)M24F-B | − | |
| 29 | 24E3; 25A7 | Df(2L)sc19-3 | − | |
| 30 | 24E4; 25B2 | Df(2L)dp-h25 | − | |
| 31 | 24F1–2; 25C5 | Df(2L)sc19-6 | − | |
| 32 | 25A5; 25D6 | Df(2L)sc19-5 | ++ | Region 3 |
| 33 | 25D2–3; 26B2–5 | Df(2L)cl-h3 | − | |
| 34 | 25F3–26A1; 26D3–11 | Df(2L)E110 | − | |
| 35 | 26D3–E1; 26F4–7 | Df(2L)BSC6 | − | |
| 36 | 26D10–E1; 27C1 | Df(2L)BSC7 | ++ | Region 4 |
| 37 | 27B2; 27F1–2 | Df(2L)spd-j2 | − | |
| 38 | 27C5–9; 28B3–4 | Df(2L)J-H | − | |
| 39 | 27E; 28C1–4 | Df(2L)spd | + | FP6 |
| 40 | 27E2; 28D1 | Df(2L)XE-3801 | +++ | FP4 |
| 41 | 28B2; 28D1 | Df(2L)XE-2750 | + | FP6 |
| 42 | 28DE (within) | Df(2L)Trf-C6R31 | ++ | FP4 |
| 43 | 28E4–7; 29B2–C1 | Df(2L)TE29Aa-11 | − | |
| 44 | 29C1–2; 30C8–9 | Df(2L)N22-14 | ++ | FP4 |
| 45 | 29C3–5; 30C8–9 | Df(2L)N22-5 | ++ | Regions 5, 6 |
| 46 | 29E2–F1; 30C2–4 | Df(2L)TE30Cb-1 | − | |
| 47 | 30A1–2; 30D1–2 | Df(2L)N22-3 | ++ | Region 6 |
| 48 | 30A3–5; 30C5 | Df(2L)30A-C | ++ | Region 6 |
| 49 | 30A9–B1; 30D2–F4 | Df(2L)gamma7 | +++ | FP4 |
| 50 | 30D1–F6; 31F1–5 | Df(2L)Mdh | − | |
| 51 | 31B1; 32A1–2 | Df(2L)J2 | − | |
| 52 | 31D1–11; 31E7 | Df(2L)J27 | − | |
| 53 | 32D1; 32F1–3 | Df(2L)FCK-20 | − | |
| 54 | 32F1–3; 33F1–2 | Df(2L)Prl | + | FP5, -6 |
| 55 | 33A1; 33B1–2 | Df(2L)esc-P2-0 | + | FP6 |
| 56 | 33A1; 33B2 | Df(2L)esc10 | − | |
| 57 | 33A1; 33E | Df(2L)esc-P3-0 | − | |
| 58 | 33B3; 34A1–2 | Df(2L)prd1.7 | − | |
| 59 | 34B7–12; 34E3 | In(2L)b82a1 | ++ | Region 7 |
| 60 | 34C1; 35C1 | Df(2L)b87e25 | ++ | FP2, -5 |
| 61 | 34C4; 35A4 | Df(2L)b80e3 | − | |
| 62 | 34D1–2; 35C1 | Df(2L)64j | +++ | FP1 |
| 63 | 34D3–4; 35C1 | Df(2L)fn30 | ++++ | FP1 |
| 64 | 34D4; 34E3 | Df(2L)b88f32 | ++ | FP1 |
| 65 | 34E2; 35B3–4 | Df(2L)fn7 | +++ | FP1 |
| 66 | 34E3; 35D2–5 | Df(2L)el80f1 | + | FP1, -6 |
| 67 | 34E4–34F1; 35C3 | Df(2L)noc11 | − | |
| 68 | 34E5–F1; 35C3 | Df(2L)A263 | − | |
| 69 | 34F2–5; 35C4 | Df(2L)osp38 | ++ | FP1 |
| 70 | 34F4; 35C3 | Df(2L)fn5 | − | |
| 71 | 34F4–5; 35D4–5 | Df(2L)fn1 | ++++ | FP1 |
| 72 | 34F5; 35B2 | Df(2L)el81i1 | − | |
| 73 | 34F5; 35B10 | Df(2L)TE35BC-31 | − | |
| 74 | 34F5–35A4; 35D2 | Df(2L)do1 | − | |
| 75 | 35A1–4; 35C1–3 | Df(2L)A400 | − | |
| 76 | 35A4–B1; 35B2 | Df(2L)fn2 | + | FP1 |
| 77 | 35B1; 35F1 | Df(2L)A446 | − | |
| 78 | 35B3; 35E6 | Df(2L)osp29 | − | |
| 79 | 35B4–6; 35E1–2 | Df(2L)TE35BC-24 | − | |
| 80 | 35D1; 36A6–7 | Df(2L)r10 | ++ | Region 8 |
| 81 | 35F6–12; 36D | Df(2L)cact-255rv64 | +++ | FP5 |
| 82 | 36A8–9; 36F1 | Df(2L)H20 | − | |
| 83 | 36C2–4; 37B9–10 | Df(2L)TW137 | − | |
| 84 | 36E4–F1; 38A6–7 | Df(2L)TW50 | +++ | FP1 |
| 85 | 36F7–9; 37B2–7 | Df(2L)TW3 | − | |
| 86 | 36F7–9; 37D1–2 | Df(2L)VA16 | ++ | FP1 |
| 87 | 37B2–8; 37C5 | Df(2L)hk-UC2 | − | |
| 88 | 37B2–10; 38D2–5 | Df(2L)pr-A16 | − | |
| 89 | 37B2–8; 37E2 | Df(2L)TW158 | ++ | FP1 |
| 90 | 37B9–10; 37D1–2 | Df(2L)TW130 | ++ | FP1 |
| 91 | 37B9–10; 37D5 | Df(2L)VA23 | ++ | FP1 |
| 92 | 37C1; 37F5 | Df(2L)VA17 | ++ | FP1 |
| 93 | 37C2–5; 38B2–C1 | Df(2L)VA12 | +++ | FP1 |
| 94 | 37C2–7; 38C1–2 | Df(2L)Sd77 | − | |
| 95 | 37D2; 38A1 | Df(2L)E55 | +++ | FP1 |
| 96 | 37D2–5; 38A6–B2 | Df(2L)Sd37 | − | |
| 97 | 37D2–5; 39A4–7 | Df(2L)pr-A14 | ++ | FP1 |
| 98 | 37D6–E1; 38E6–9 | Df(2L)TW2 | ++ | FP1 |
| 99 | 37E2–4; 39D1 | Df(2L)TW12 | − | |
| 100 | 37E2–F1; 38B5–C1 | Df(2L)TW9 | +++ | FP1 |
| 101 | 38A1; 39D3–E1 | Df(2L)TW84 | + | FP1, -6 |
| 102 | 38A1; 39F1 | Df(2L)TW65 | ++ | Region 9 |
| 103 | 38A3–4; 38B6–C1 | Df(2L)pr-A20 | ++ | FP1 |
| 104 | 38A7–B1; 39C2–3 | Df(2L)TW1 | ++ | FP1 |
| 105 | 38B3–6; 40A3 | Df(2L)pr-M1 | + | FP6 |
| 106 | 38E2; 39E7 | Df(2L)DS6 | − | |
| 107 | 40h35; 40h38L | Df(2L)C' | − | |
| 108 | h38R; h46 | Df(2R)M41A10 | + | FP6 |
| 109 | h42–h43; 42A2–3 | In(2R)bwVDe2LCyR | + | FP6 |
| 110 | h44–h46; 41B1–41F11 | Df(2R)M41A8 | +++ | FP4, -5 |
| 111 | h44–h46; 42A1–2 | Df(2R)M41A4 | ++ | FP4 |
| 112 | 41BC; 42A16–B1 | Df(2R)nap14 | + | FP6 |
| 113 | 41D2–E1; 42B1–3 | Df(2R)nap1 | − | |
| 114 | 41F3–4; 42A3–9 | Df(2R)17I | ++ | FP1 |
| 115 | 42A1–2; 42E6–F1 | Df(2R)nap9 | − | |
| 116 | 42A1–19; 42E2–7 | Df(2R)cn88b | − | |
| 117 | 42B3–4; 43E18 | Df(2R)ST1 | ++ | FP1 |
| 118 | 42B4–C1; 43F–44A1 | Df(2R)cn87e | − | |
| 119 | 42C1–7; 43F5–8 | Df(2R)pk78s | − | |
| 120 | 42C2; 42D2–3 | Df(2R)42 | − | |
| 121 | 42C2–7; 43D1–7 | Df(2R)Drl-rv17 | + | FP1, -6 |
| 122 | 42E; 44C1 | Df(2R)cn9 | ++ | FP1 |
| 123 | 42E1–4; 43C3 | Df(2R)Drl-rv3 | ++ | FP1 |
| 124 | 43A3; 43F6 | Df(2R)P32a | + | FP1, -6 |
| 125 | 43C5; 44B6–C1 | Df(2R)cn83c | + | FP1, -6 |
| 126 | 43C7; 43F2–8 | Df(2R)cn-S6 | − | |
| 127 | 43E7–18; 44B4–5 | Df(2R)CA53 | − | |
| 128 | 43F; 44D3–8 | Df(2R)H3C1 | − | |
| 129 | 44C1–2; 44E1–4 | Df(2R)44CE | − | |
| 130 | 44D1–4; 44F12 | Df(2R)H3E1 | − | |
| 131 | 44D2–E1; 45B8–C1 | Df(2R)Np3 | − | |
| 132 | 44F11; 45C1 | Df(2R)Np4 | ++ | FP1 |
| 133 | 44F11; 45D9–E1 | Df(2R)Np5 | + | FP1, -6 |
| 134 | 44F2–3; 45C6 | Df(2R)Np1 | − | |
| 135 | 45A6–7; 45E2–3 | Df(2R)w45-30n | + | FP4 |
| 136 | 45A9–10; 45D5–8 | Df(2R)w73-1 | − | |
| 137 | 45C8; 45D8 | Df(2R)wun-GL | ++ | FP1 |
| 138 | 45C8–D10; 45D9–E1 | Df(2R)w45-19g | ++ | FP1 |
| 139 | 45D3–4; 45F2–6 | Df(2R)BSC29 | − | |
| 140 | 46A1–4; 46C3–12 | Df(2R)B5 | + | FP6 |
| 141 | 46C1–2; 46E1–2 | Df(2R)X3 | − | |
| 142 | 46C2; 47A1 | Df(2R)X1 | − | |
| 143 | 46C3–4; 46C9–11 | Df(2R)eve | +++ | FP1, -2 |
| 144 | 46F1; 47A10 | Df(2R)12 | ++ | FP2 |
| 145 | 46F1; 47B9 | Df(2R)stan2 | ++ | FP3 |
| 146 | 47A3; 47E | Df(2R)E3363 | − | |
| 147 | 47D3; 48A5 | Df(2R)en-A | + | FP6 |
| 148 | 47E3; 48A5–B2 | Df(2R)en-B | ++ | FP2, -4 |
| 149 | 48A1; 48B5 | Df(2R)en-SFX31 | +++ | FP3 |
| 150 | 48A1–2; 48B–C1 | Df(2R)en28 | + | FP3, -6 |
| 151 | 48A3; 48C6–8 | Df(2R)en30 | − | |
| 152 | 48C5–D1; 48D5–E11 | Df(2R)BSC39 | − | |
| 153 | 48E; 49A | Df(2R)CB21 | +++ | FP2, -5 |
| 154 | 49A; 49E1–2 | Df(2R)vg135 | + | FP5, -6 |
| 155 | 49B2–3; 49E2 | Df(2R)vg-C | − | |
| 156 | 49C1–2; 49E6 | Df(2R)vg-D | ++ | Region 10 |
| 157 | 49C1–4; 50C23–D1 | Df(2R)CX1 | +++ | FP4 |
| 158 | 49D3–4; 50A2 | Df(2R)vg-B | + | FP6 |
| 159 | 50C21–23; 50D1–5 | Df(2R)50C-101 | − | |
| 160 | 50E6–F1; 51E2–4 | Df(2R)BSC11 | +++ | FP4 |
| 161 | 50F6–9; 51B3 | Df(2R)L48 | − | |
| 162 | 51A2; 51B6 | Df(2R)trix | ++ | FP1 |
| 163 | 51A5; 51C1 | Df(2R)03072 | − | |
| 164 | 51C3–7; 51E7–11 | Df(2R)14 | − | |
| 165 | 51D1–2; 51E5 | Df(2R)XTE-58 | − | |
| 166 | 51D3–E1; 52D1 | Df(2R)XTE-18 | − | |
| 167 | 51F13; 52F8–9 | Df(2R)Jp4 | +++ | Region 11 |
| 168 | 52A13–14; 52F10–11 | Df(2R)Jp5 | +++ | Region 11 |
| 169 | 52A9–10; 52D9–15 | Df(2R)WMG | − | |
| 170 | 52D3; 53A1 | Df(2R)Jp6 | ++ | Region 11 |
| 171 | 54E8–F1; 55B9–C1 | Df(2R)Pcl7B | − | |
| 172 | 54F2; 56A1 | Df(2R)RM2-1 | +++ | FP3 |
| 173 | 55A1; 55C1–3 | Df(2R)Pcl11B | − | |
| 174 | 55A1; 55F1–2 | Df(2R)PC4 | − | |
| 175 | 55C1–2; 56B1–2 | Df(2R)C29 | ++ | FP3 |
| 176 | 55D2–E1; 56B2 | Df(2R)PC66 | ++ | FP3 |
| 177 | 55E6–F3; 56C1 | Df(2R)P34 | ++ | FP2 |
| 178 | 56D7–E3; 56F9–12 | Df(2R)BSC22 | + | FP6 |
| 179 | 56F 5; 56F15 | Df(2R)173 | − | |
| 180 | 56F 5; 56F15 | Df(2R)017 | − | |
| 181 | 56F 9–11; 57D12 | Df(2R)AA21 | + | FP3, -6 |
| 182 | 56F12–14; 57A4 | Df(2R)BSC19 | − | |
| 183 | 57A3; 57B1 | Df(2R)exu2 | +++ | FP2 |
| 184 | 57A6; 57B6 | Df(2R)D4 | − | |
| 185 | 57B1; 57B13–14 | Df(2R)E2 | − | |
| 186 | 57B4; 58B1–2 | Df(2R)Pu-D17 | − | |
| 187 | 57D2–8; 58D1 | Df(2R)Egfr5 | − | |
| 188 | 58B1–2; 58E4 | Df(2R)X58-7 | − | |
| 189 | 58B3; 59A1 | Df(2R)X58-8 | − | |
| 190 | 58C3–7; 58D6–8 | Df(2R)X58-3 | − | |
| 191 | 58D1–2; 59A1 | Df(2R)X58-12 | +++ | Region 12 |
| 192 | 59A1–3; 59B1–2 | Df(2R)59AB | ++ | Region 12 |
| 193 | 59A1–3; 59D1–4 | Df(2R)59AD | − | |
| 194 | 59D 4–8; 59D9–E1 | Df(2R)vir-12 | − | |
| 195 | 59D 8; 60A7 | Df(2R)bw-S46 | + | FP6 |
| 196 | 59D11; 59F6–8 | Df(2R)bw-HB132 | + | FP4 |
| 197 | 59E; 60A1 | Df(2R)egl2 | ++ | FP1 |
| 198 | 59E1; 59F6 | Df(2R)bw5 | + | FP6 |
| 199 | 59E1; 60C7–D1 | Df(2R)bwVDe2LPxKR | − | |
| 200 | 59F1; 59F5 | Df(2R)egl3 | ++ | FP1 |
| 201 | 59F3; 60A8–16 | Df(2R)G10-7-5 | − | |
| 202 | 59F6; 60A12–16 | Df(2R)or-BR11 | − | |
| 203 | 60B8–10; 60D1 | Df(2R)Px1 | − | |
| 204 | 60C6; 60D9–10 | Df(2R)Px2 | − | |
| 205 | 60E6; 60F1–2 | Df(2R)ES1 | ++ | FP2, -4 |
| 206 | 60E1–2; 60E6 | Df(2R)Dll-MP | ++ | FP4 |
| 207 | 60E10; 60F5 | Df(2R)Kr10 | ++ | FP4 |
| 208 | 60E6–9; 60E11 | Df(2R)M60E | − | |
| 209 | 60E9; 60F1 | Df(2R)gsb | + | FP3, -6 |
| 210 | 60F2; 60F5 | Df(2R)Kr14 | − | |
| 211 | 61A; 61D3 | Df(3L)emc-E12 | − | |
| 212 | 61A1; 61B | Df(3L)B71 | − | |
| 213 | 61C1–4; 61F3 | Df(3L)Ar12-1 | + | FP6 |
| 214 | 61C3–4; 61E | Df(3L)Ar11 | + | FP6 |
| 215 | 61C4; 62A8 | Df(3L)Ar14-8 | + | FP6 |
| 216 | 61D3–E1; 61F5–8 | Df(3L)bab-PG | − | |
| 217 | 61F8; 62A3–5 | Df(3L)ru-22 | + | FP6 |
| 218 | 62A10–B1; 62C4–D1 | Df(3L)R-G5 | − | |
| 219 | 62A10–B1; 62D2 | Df(3L)Aprt-1 | − | |
| 220 | 62B9; 62E7 | Df(3L)R-G7 | − | |
| 221 | 63C1; 63D3 | Df(3L)HR232 | − | |
| 222 | 63C1–2; 63F1–2 | Df(3L)1227 | ++ | FP1 |
| 223 | 63C6; 63F7 | Df(3L)HR119 | − | |
| 224 | 63E2; 64B17 | Df(3L)GN50 | − | |
| 225 | 63F6–7; 64C13–15 | Df(3L)GN24 | − | |
| 226 | 64; 65B5–C1 | Dr(3L)CH39 | − | |
| 227 | 64B–C; 65B5–C1 | Df(3L)CH18 | − | |
| 228 | 64C; 65C | Df(3L)ZN47 | − | |
| 229 | 64E1–13; 65C1–D6 | Df(3L)v65c | ++ | FP1 |
| 230 | 65A; 65E1 | Df(3L)W5.4 | − | |
| 231 | 65A2; 65E1 | Df(3L)XDl98 | − | |
| 232 | 65D4–5; 65E4–6 | Df(3L)BSC27 | − | |
| 233 | 65E1–12; 66A17 | Df(3L)RM5-2 | − | |
| 234 | 65F3; 66B10 | Df(3L)pbl-X1 | − | |
| 235 | 66A17–20; 66C1–5 | Df(3L)ZP1 | − | |
| 236 | 66B12–C1; 66D2–4 | Df(3L)BSC13 | ++ | Region 13 |
| 237 | 66B8–9; 66C9–10 | Df(3L)66C-G28 | +++ | Region 13 |
| 238 | 66C7–10; 66C7–10 | Df(3L)66C-I65 | − | |
| 239 | 66E1–6; 66F1–6 | Df(3L)Scf-R6 | − | |
| 240 | 66E3–4; 66F1–2 | Df(3L)Scf-R11 | − | |
| 241 | 66F5; 66F5 | Df(3L)Rdl-2 | − | |
| 242 | 66F5; 67B1 | Df(3L)29A6 | ++ | Region 14 |
| 243 | 67A2; 67D13 | Df(3L)AC1 | + | FP6 |
| 244 | 67E1–2; 68C1–2 | Df(3L)lxd6 | − | |
| 245 | 68A2; 69A1 | Df(3L)vin5 | − | |
| 246 | 68C8; 69B4–5 | Df(3L)vin7 | − | |
| 247 | 69A4–5; 69D4–6 | Df(3L)eyg-C1 | − | |
| 248 | 69B1–5; 69D1–6 | Df(3L)iro-2 | − | |
| 249 | 69D; 69D | Df(3L)8ex25 | + | FP1, -6 |
| 250 | 69D2; 69E3–5 | Df(3L)E44 | − | |
| 251 | 69D4–5; 69F5–7 | Df(3L)BSC10 | − | |
| 252 | 69F3–4; 70C3–4 | Df(3L)C190LUbx42TR | − | |
| 253 | 70C2; 72A1 | Df(3L)D-5rv12 | − | |
| 254 | 70C2–6; 70E1 | Df(3L)fz-CAL | − | |
| 255 | 70D2; 70E8 | Df(3L)fz-D21 | − | |
| 256 | 70D2; 71E4–5 | Df(3L)fz-M21 | − | |
| 257 | 70E; 71F | Df(3L)Brd6 | ++ | FP1 |
| 258 | 71A1–2; 71C1–2 | Df(3L)Brd15 | − | |
| 259 | 71C2–3; 72B1–C1 | Df(3L)XG-5 | − | |
| 260 | 71C3; 71E5 | Df(3L)BK10 | − | |
| 261 | 72A2; 72D10 | Df(3L)th102 | − | |
| 262 | 72A3–4; 72D1–5 | Df(3L)brm11 | − | |
| 263 | 72C1; 73A4 | Df(3L)st-f13 | − | |
| 264 | 72D10–11; 73D1–2 | Df(3L)st-b11 | − | |
| 265 | 73A3; 74F1–4 | Df(3L)81k19 | − | |
| 266 | 74D3–75A1; 75B2–5 | Df(3L)BSC8 | − | |
| 267 | 75B10; 75C5 | Df(3L)W4 | − | |
| 268 | 75C1–2; 75F1 | Df(3L)Cat | − | |
| 269 | 75F10–11; 76A1–5 | Df(3L)fz2 | − | |
| 270 | 76A7–B1; 76B4–5 | Df(3L)BSC20 | − | |
| 271 | 76B; 76F | Df(3L)XS2182 | − | |
| 272 | 76B; 77A | Df(3L)XS543 | − | |
| 273 | 76B1–2; 76D5 | Df(3L)kto2 | ++ | FP1 |
| 274 | 76B4; 76D3 | Df(3L)XS705 | − | |
| 275 | 76B4; 77B | Df(3L)XS-533 | − | |
| 276 | 76B6; 77C1 | Df(3L)XS572 | − | |
| 277 | 77A1; 77D1 | Df(3L)rdgC-co2 | − | |
| 278 | 77B7–9; 77F1–5 | Df(3L)ri-79c | ++ | Region 15 |
| 279 | 77E2; 78A4 | Df(3L)ri-XT1 | − | |
| 280 | 77F3; 78C8–9 | Df(3L)ME107 | − | |
| 281 | 78C5–6; 78E3–79A1 | Df(3L)Pc-2q | − | |
| 282 | 79C; 79E1–8 | Df(3L)Ten-m-AL29 | − | |
| 283 | 79D3–E1; 79F3–6 | Df(3L)HD1 | − | |
| 284 | 79E1–2; 79E1–8 | Df(3L)Ten-m-AL1 | − | |
| 285 | 79E5–F1; 80A2–3 | Df(3L)BSC21 | − | |
| 286 | 79F; 80A | Df(3L)Delta1AK | − | |
| 287 | 80Fb–g | Df(3L)3-52 | + | FP6 |
| 288 | 80Fd–e | Df(3R)6-61 | + | FP6 |
| 289 | 80Ff–g | Df(3L)8A-80 | − | |
| 290 | 80Fg–j | Df(3L)1-166 | − | |
| 291 | 80Fh–j | Df(3L)2-66 | + | FP6 |
| 292 | 80Fj | Df(3L)2-30 | + | FP6 |
| 293 | 81 Fab | Df(3R)4-75 | − | |
| 294 | 81F; 82F10–11 | Df(3R)2-2 | − | |
| 295 | 81F3–6; 82F5–7 | Df(3R)ME15 | + | FP6 |
| 296 | 82A5–6; 82E4 | Df(3R)Z1 | − | |
| 297 | 82C4; 82F3 | Df(3R)110 | − | |
| 298 | 82D5; 82F3–6 | Df(3R)6-7 | + | FP1, -6 |
| 299 | 82F3–4; 82F10–11 | Df(3R)3-4 | − | |
| 300 | 82F8–10; 83A1–3 | Df(3R)e1025-14 | − | |
| 301 | 83B7–C1; 83C6–D1 | Df(3R)BSC47 | − | |
| 302 | 83E1–2; 84B1 | Df(3R)WIN11 | ++ | Region 16 |
| 303 | 83E3; 84B1 | Df(3R)Dfd13 | +++ | Region 16 |
| 304 | 84A1; 84B1 | Df(3R)9A99 | − | |
| 305 | 84A1–2; 84B1–2 | Df(3R)Scr | − | |
| 306 | 84A6–B1; 84D4–D9 | Df(3R)roe | − | |
| 307 | 84C1–2; 84E1 | Df(3R)dsx2M | − | |
| 308 | 84C8; 84F6 | Df(3R)dsx29 | − | |
| 309 | 84D 4–6; 85B6 | Df(3R)p712 | ++ | FP1 |
| 310 | 84D 8; 85B3–5 | Df(3R)dsx37 | − | |
| 311 | 84D 8–9; 85A1–2 | Df(3R)dsx11 | − | |
| 312 | 84D11; 84E8 | Df(3R)dsx15 | − | |
| 313 | 84E8–9; 85B6 | Df(3R)p40 | ++ | FP1 |
| 314 | 84F1; 85A6–B9 | Df(3R)p13 | − | |
| 315 | 84F2; 85A5–7 | Df(3R)CA3 | ++ | FP1 |
| 316 | 85A2; 85C1–2 | Df(3R)p-XT103 | − | |
| 317 | 85D8; 85E10–13 | Df(3R)by10 | − | |
| 318 | 85D11–13; 85F6 | Df(3R)by62 | +++ | Region 17 |
| 319 | 85D12; 85E10 | Df(3R)GB104 | − | |
| 320 | 86C1; 87B5 | Df(3R)M-Kx1 | − | |
| 321 | 86E2–3; 87C6–7 | Df(3R)T-32 | − | |
| 322 | 86F1–2; 87C6–7 | Df(3R)T-10 | ++ | FP1 |
| 323 | 87B12; 87E8 | Df(3R)ry615 | − | |
| 324 | 87D2; 87F2 | Df(3R)ry27 | − | |
| 325 | 87E–F; 88B | Df(3R)CbxTwtLUbxKM5R | + | FP1, -6 |
| 326 | 87E1; 87F11 | Df(3R)I26c | − | |
| 327 | 87F1; 87F15 | Df(3R)urd | − | |
| 328 | 87F12–14; 88C2 | Df(3R)red31 | − | |
| 329 | 88A2; 88C1–D1 | Df(3R)red1 | − | |
| 330 | 88B1; 88C2 | Df(3R)red-P93 | − | |
| 331 | 88E7–13; 89A1 | Df(3R)ea | + | FP6 |
| 332 | 88F7; 89A11–13 | Df(3R)Po4 | − | |
| 333 | 89A1–2; 89A11–13 | Df(3R)Po2 | ++ | FP1 |
| 334 | 89A1–2; 89A11–13 | Df(3R)Po3 | − | |
| 335 | 89A8; 89B3 | Df(3R)Exel7327 | − | |
| 336 | 89B4; 89B10 | Df(3R)sbd45 | − | |
| 337 | 89B5; 89C | Dr(3R)sd104 | − | |
| 338 | 89B5–6; 89E2–3 | Df(3R)bxd100 | + | FP1, -6 |
| 339 | 89B7–8; 89E7–8 | Df(3R)P115 | − | |
| 340 | 89E1–89F4; 91B1–B2 | Df(3R)DG2 | − | |
| 341 | 89E2–3; 90A | Df(3R)C4 | ++ | FP1 |
| 342 | 89E2–3; 90D | Df(3R)RD31 | − | |
| 343 | 90F1–2; 91F5 | Df(3R)Cha7 | − | |
| 344 | 91A2–B3; 91F13–92A1 | Df(3R)Cha1a | − | |
| 345 | 91F1–2; 92D3–6 | Df(3R)D1-BX12 | − | |
| 346 | 92B3; 92F13 | Df(3R)H-B79 | − | |
| 347 | 93B2–13; 94A3–8 | Df(3R)e-N19 | − | |
| 348 | 93B6; 93D3–4 | Df(3R)e-R1 | − | |
| 349 | 93C6; 94A1–4 | Df(3R)e-GC3 | + | FP1, -6 |
| 350 | 93E–F; 94C–D | Df(3R)5C1 | + | FP1, -6 |
| 351 | 93F11–14; 94D10–13 | Df(3R)hh | − | |
| 352 | 95A5–7; 95C10–11 | Df(3R)mbc-30 | ++ | Region 18 |
| 353 | 95A5–7; 95D6–11 | Df(3R)mbc-R1 | − | |
| 354 | 95D7–11; 95F15 | Df(3R)crb-F89-4 | − | |
| 355 | 95D11–E2; 96A2 | Df(3R)crb87-4 | − | |
| 356 | 96A1; 96A20–25 | Df(3R)Ubx7LLatsR | − | |
| 357 | 96A2–7; 96D2–4 | Df(3R)slo8 | − | |
| 358 | 96F1; 97B1 | Df(3R)Espl3 | − | |
| 359 | 96F10–11; 96F11 | Df(3R)Espl22 | − | |
| 360 | 96F12–14; 97C4–5 | Df(3R)ME61 | − | |
| 361 | 97A; 98A1–2 | Df(3R)T1-P | − | |
| 362 | 97E3; 98A5 | Df(3R)D605 | − | |
| 363 | 98D3–7; 98D3–7 | Df(3R)M15 | − | |
| 364 | 98E3; 99A6 | Df(3R)3450 | − | |
| 365 | 98F14; 99E2–3 | Df(3R)R133 | − | |
| 366 | 99A6; 99C1 | Df(3R)01215 | − | |
| 367 | 99D1–2; 99E1 | Df(3R)X3F | − | |
| 368 | 99F1–2; 100B5 | Df(3R)tll-g | − | |
| 369 | 100A2; 100C2–3 | Df(3R)tll-e | + | FP6 |
| 370 | 100D1; 100D3–4 | Df(3R)awd-KRB | − | |
| 371 | 100D1–2; 100E–F | Df(3R)faf-BP | − | |
| 372 | 100D2; 100F5 | Df(3R)04661 | − |
Breakpoints are as determined by FlyBaseConsortium (2003); otherwise breakpoints were supplied by the Bloomington stock center.
−, nonsuppressor; +, equivocal suppressor with a phenotype that overlaps the nonsuppressed phenotype; ++, weak suppressor; +++, moderate suppressor; ++++, strong suppressor (see Figure 1).
Determination of suppressor on the deficiency chromosome. FP1, false positive because the region of the deficiency is covered by one or more nonsuppressing deficiencies; FP2, false positive because the suppressor maps to the tip of 2L rather than to the site of the deficiency; FP3, false positive because the region of the deficiency is covered by a combination of nonsuppressing deficiencies and deficiencies with suppressors that map to the 2L tip; FP4, assumed false positive because the 2L TAS is missing by in situ hybridization; FP5, assumed false positive because the deficiency chromosome fails to complement lethal mutations of l(2)gl; FP6, the suppressor cannot be adequately tested because the phenotype overlaps wild type. Regions refer to map positions as shown in Figures 3 and 4.
. As the P{wvar}KR3-2 insert carries a w+ telomeric transgene, thus necessitating a null white allele on the X chromosome, it was not possible to test most X chromosome deficiencies in this assay. We, therefore, tested only autosomal deficiencies. In the discussion below, deficiencies are referred to by their sequence numbers in Table 1.
To start, the standard deficiency kits were used to screen the maximum fraction of the genome with the minimum number of deficiencies. We assumed that all null alleles would have the same phenotype, and thus deficiencies on chromosomes that do not have a suppressor phenotype identify regions devoid of dosage-sensitive suppressor genes. As regions of potential interest were identified, additional deficiencies were obtained to verify and refine the position of a potential suppressor. Within the limits of the stock center collection, we tested deficiencies for any given locus until we found a chromosome that did not have a suppressor phenotype. This led to unequal coverage of the genome, with some regions tested several times.
Some of the deficiencies could not be tested. Thirty deficiency chromosomes carried cryptic white genes that became obvious only in a control cross to y w67c23; + females that was run in parallel with the test cross. Twenty-two others required a duplication for viability. Six stocks were insufficiently marked to allow us to easily distinguish the deficiency from the balancer chromosome. In total, we tested 372 deficiency chromosomes for suppression of TPE, 210 for chromosome 2, and 162 for chromosome 3.
As chromosomes were tested, the deficiencies were aligned on a cytogenetic map to identify sites of potential suppressor genes. With the assumption that deficiencies with a nonsuppressor phenotype identified regions devoid of suppressor genes, it quickly became obvious that many of the tested chromosomes carried suppressors that are not within the bounds of the deficiencies being tested. These were deemed to be false positive results (Table 1; FP1). With a high frequency of false positives (58/149 = 0.39), it became necessary to verify the suppressors on as many of the deficiency chromosomes as possible. Therefore, all second chromosome deficiencies with suppressor phenotypes were chosen for further analysis, but we could obtain data on only 40 (Table 2)
Tests to distinguish whether a suppressor is at the 2L telomere or the named deficiency
Df no.a . | Genetic Map Positionb . | 2L TAS . | l(2)gl lethal complementation . | Callc . |
|---|---|---|---|---|
| 7 | — | Weak | Viable | FP4 |
| 8 | — | No | Lethal | FP4, -5 |
| 20 | — | No | Viable | FP1, -4 |
| 21 | — | Yes | Viable | Region 2 |
| 24 | 0.4 (256) | No | Viable | FP2, -4 |
| 36 | — | Yes | Viable | Region 4 |
| 40 | — | Weak | Viable | FP4 |
| 42 | — | Weak | Viable | FP4 |
| 44 | — | Weak | Viable | FP4 |
| 48 | — | Yes | Viable | Region 6 |
| 49 | — | No | Viable | FP4 |
| 59 | — | Yes | Viable | Region 7 |
| 60 | 0.2 (442) | Yes | Lethal | FP2, -5 |
| 80 | — | Yes | Viable | Region 8 |
| 81 | — | — | Lethal | FP5 |
| 102 | — | Yes | Viable | Region 9 |
| 110 | — | No | Lethal | FP4, -5 |
| 111 | — | Weak | Viable | FP4 |
| 135 | — | Weak | — | FP4 |
| 137 | — | Yes | Viable | FP1 |
| 138 | — | Yes | Viable | FP1 |
| 143 | 0.4 (382) | — | Viable | FP1, -2 |
| 144 | 0.4 (145) | Yes | Viable | FP2 |
| 145 | — | Yes | Viable | FP3 |
| 148 | 0.4 (210) | Weak | Viable | FP2, -4 |
| 149 | — | Yes | — | FP3 |
| 153 | 0.4 (674) | — | Lethal | FP2, -5 |
| 157 | — | Weak | Viable | FP4 |
| 160 | — | No | Viable | FP4 |
| 167 | — | Yes | Viable | Region 11 |
| 172 | — | Yes | Viable | FP3 |
| 176 | — | Yes | Viable | FP3 |
| 177 | −0.1 (350) | Yes | Viable | FP2 |
| 183 | 0.1 (336) | — | Viable | FP2 |
| 191 | — | Yes | Viable | Region 12 |
| 192 | — | Yes | Viable | Region 12 |
| 196 | — | Weak | — | FP4 |
| 205 | 0.4 (332) | No | Viable | FP2, -4 |
| 206 | — | No | Viable | FP4 |
| 207 | — | Weak | Viable | FP4 |
Df no.a . | Genetic Map Positionb . | 2L TAS . | l(2)gl lethal complementation . | Callc . |
|---|---|---|---|---|
| 7 | — | Weak | Viable | FP4 |
| 8 | — | No | Lethal | FP4, -5 |
| 20 | — | No | Viable | FP1, -4 |
| 21 | — | Yes | Viable | Region 2 |
| 24 | 0.4 (256) | No | Viable | FP2, -4 |
| 36 | — | Yes | Viable | Region 4 |
| 40 | — | Weak | Viable | FP4 |
| 42 | — | Weak | Viable | FP4 |
| 44 | — | Weak | Viable | FP4 |
| 48 | — | Yes | Viable | Region 6 |
| 49 | — | No | Viable | FP4 |
| 59 | — | Yes | Viable | Region 7 |
| 60 | 0.2 (442) | Yes | Lethal | FP2, -5 |
| 80 | — | Yes | Viable | Region 8 |
| 81 | — | — | Lethal | FP5 |
| 102 | — | Yes | Viable | Region 9 |
| 110 | — | No | Lethal | FP4, -5 |
| 111 | — | Weak | Viable | FP4 |
| 135 | — | Weak | — | FP4 |
| 137 | — | Yes | Viable | FP1 |
| 138 | — | Yes | Viable | FP1 |
| 143 | 0.4 (382) | — | Viable | FP1, -2 |
| 144 | 0.4 (145) | Yes | Viable | FP2 |
| 145 | — | Yes | Viable | FP3 |
| 148 | 0.4 (210) | Weak | Viable | FP2, -4 |
| 149 | — | Yes | — | FP3 |
| 153 | 0.4 (674) | — | Lethal | FP2, -5 |
| 157 | — | Weak | Viable | FP4 |
| 160 | — | No | Viable | FP4 |
| 167 | — | Yes | Viable | Region 11 |
| 172 | — | Yes | Viable | FP3 |
| 176 | — | Yes | Viable | FP3 |
| 177 | −0.1 (350) | Yes | Viable | FP2 |
| 183 | 0.1 (336) | — | Viable | FP2 |
| 191 | — | Yes | Viable | Region 12 |
| 192 | — | Yes | Viable | Region 12 |
| 196 | — | Weak | — | FP4 |
| 205 | 0.4 (332) | No | Viable | FP2, -4 |
| 206 | — | No | Viable | FP4 |
| 207 | — | Weak | Viable | FP4 |
Tests to distinguish whether a suppressor is at the 2L telomere or the named deficiency
Df no.a . | Genetic Map Positionb . | 2L TAS . | l(2)gl lethal complementation . | Callc . |
|---|---|---|---|---|
| 7 | — | Weak | Viable | FP4 |
| 8 | — | No | Lethal | FP4, -5 |
| 20 | — | No | Viable | FP1, -4 |
| 21 | — | Yes | Viable | Region 2 |
| 24 | 0.4 (256) | No | Viable | FP2, -4 |
| 36 | — | Yes | Viable | Region 4 |
| 40 | — | Weak | Viable | FP4 |
| 42 | — | Weak | Viable | FP4 |
| 44 | — | Weak | Viable | FP4 |
| 48 | — | Yes | Viable | Region 6 |
| 49 | — | No | Viable | FP4 |
| 59 | — | Yes | Viable | Region 7 |
| 60 | 0.2 (442) | Yes | Lethal | FP2, -5 |
| 80 | — | Yes | Viable | Region 8 |
| 81 | — | — | Lethal | FP5 |
| 102 | — | Yes | Viable | Region 9 |
| 110 | — | No | Lethal | FP4, -5 |
| 111 | — | Weak | Viable | FP4 |
| 135 | — | Weak | — | FP4 |
| 137 | — | Yes | Viable | FP1 |
| 138 | — | Yes | Viable | FP1 |
| 143 | 0.4 (382) | — | Viable | FP1, -2 |
| 144 | 0.4 (145) | Yes | Viable | FP2 |
| 145 | — | Yes | Viable | FP3 |
| 148 | 0.4 (210) | Weak | Viable | FP2, -4 |
| 149 | — | Yes | — | FP3 |
| 153 | 0.4 (674) | — | Lethal | FP2, -5 |
| 157 | — | Weak | Viable | FP4 |
| 160 | — | No | Viable | FP4 |
| 167 | — | Yes | Viable | Region 11 |
| 172 | — | Yes | Viable | FP3 |
| 176 | — | Yes | Viable | FP3 |
| 177 | −0.1 (350) | Yes | Viable | FP2 |
| 183 | 0.1 (336) | — | Viable | FP2 |
| 191 | — | Yes | Viable | Region 12 |
| 192 | — | Yes | Viable | Region 12 |
| 196 | — | Weak | — | FP4 |
| 205 | 0.4 (332) | No | Viable | FP2, -4 |
| 206 | — | No | Viable | FP4 |
| 207 | — | Weak | Viable | FP4 |
Df no.a . | Genetic Map Positionb . | 2L TAS . | l(2)gl lethal complementation . | Callc . |
|---|---|---|---|---|
| 7 | — | Weak | Viable | FP4 |
| 8 | — | No | Lethal | FP4, -5 |
| 20 | — | No | Viable | FP1, -4 |
| 21 | — | Yes | Viable | Region 2 |
| 24 | 0.4 (256) | No | Viable | FP2, -4 |
| 36 | — | Yes | Viable | Region 4 |
| 40 | — | Weak | Viable | FP4 |
| 42 | — | Weak | Viable | FP4 |
| 44 | — | Weak | Viable | FP4 |
| 48 | — | Yes | Viable | Region 6 |
| 49 | — | No | Viable | FP4 |
| 59 | — | Yes | Viable | Region 7 |
| 60 | 0.2 (442) | Yes | Lethal | FP2, -5 |
| 80 | — | Yes | Viable | Region 8 |
| 81 | — | — | Lethal | FP5 |
| 102 | — | Yes | Viable | Region 9 |
| 110 | — | No | Lethal | FP4, -5 |
| 111 | — | Weak | Viable | FP4 |
| 135 | — | Weak | — | FP4 |
| 137 | — | Yes | Viable | FP1 |
| 138 | — | Yes | Viable | FP1 |
| 143 | 0.4 (382) | — | Viable | FP1, -2 |
| 144 | 0.4 (145) | Yes | Viable | FP2 |
| 145 | — | Yes | Viable | FP3 |
| 148 | 0.4 (210) | Weak | Viable | FP2, -4 |
| 149 | — | Yes | — | FP3 |
| 153 | 0.4 (674) | — | Lethal | FP2, -5 |
| 157 | — | Weak | Viable | FP4 |
| 160 | — | No | Viable | FP4 |
| 167 | — | Yes | Viable | Region 11 |
| 172 | — | Yes | Viable | FP3 |
| 176 | — | Yes | Viable | FP3 |
| 177 | −0.1 (350) | Yes | Viable | FP2 |
| 183 | 0.1 (336) | — | Viable | FP2 |
| 191 | — | Yes | Viable | Region 12 |
| 192 | — | Yes | Viable | Region 12 |
| 196 | — | Weak | — | FP4 |
| 205 | 0.4 (332) | No | Viable | FP2, -4 |
| 206 | — | No | Viable | FP4 |
| 207 | — | Weak | Viable | FP4 |
. The primary difficulty in obtaining useful data was due to the health of the deficiency flies; many stocks had such poor viability on outcrossing that they could not be tested further.
Meiotic recombination mapping of TPE suppressors:
The most informative test, and the most demanding in terms of the health of deficiency-bearing individuals, was genetic mapping of the suppressor using a second chromosome marked with the recessive mutation al and the dominant mutations S wgSp-1 Tft nwB PinYt. This is one of the few multiply marked chromosomes at the Bloomington stock center useful for recombination studies that did not carry a suppressor of TPE. No such third chromosome could be found; thus the third chromosome suppressors were not mapped genetically. In this assay, y w67c23; Df/SM1 females were crossed to y w67c23; al S wgSp-1 Tft nwB PinYt/+ males, and Cy+ F1 multiply marked females were mated with y w67c23; P{wvar}KR3-2 al males. F2 progeny were scored for the visible markers as well as eye color.
The suppressors on nine deficiency chromosomes were mapped by meiotic recombination (Table 2). In each case the suppressor was inseparable from or to the left of al, the left-most marker, which is located at 0.4 on the genetic map. In no case was there any evidence for a suppressor at the site of the deficiency. There may be some question about the separation of the suppressor on the Df(2L)S2590 (Df 24 in Table 1) chromosome from the deficiency itself, as this deficiency removes a region in 23DE, close to the 2L tip. The suppressor, however, clearly maps to the left of S (three crossovers/256 chromosomes), while the deficiency clearly is to the right of S, which is at 21EF. Thus, all nine cases constitute false positives (Table 1, FP2). As these tests clearly separated the suppressors from the deficiencies, further regions devoid of suppressors were identified, and more deficiencies could be eliminated as causing a suppressor phenotype (Table 1, FP3).
The suppressors on most deficiency chromosomes could not be mapped genetically. Among the deficiency chromosomes with a suppressor phenotype, 44 were deemed to be neither FP1 nor FP3. Of these, 5 deficiencies are to the left of al and could not be separated from the 2L tip, 4 are too sick to attempt genetic mapping, 1 died in our lab and at the Blooming stock center and could not be tested, 16 are inviable or sterile in combination with y w67c23; SM1, 4 are inviable or sterile in combination with y w67c23; al S wgSp-1 Tft nwB PinY, 4 have suppressor phenotypes too weak to map, and 8 were mapped.
2L TAS on deficiency chromosomes:
We have shown previously (Golubovskyet al. 2001; Masonet al. 2003b) that silencing of brown-red variants of P{wvar} is suppressed by a complete, or even a partial, deficiency of the 2L TAS array on the homolog. A recent search for radiation-induced suppressors of telomeric silencing on chromosome 2 produced almost exclusively deficiencies of 2L TAS (A. Y. Konev and J. M. Mason, unpublished results). We, therefore, tested 36 deficiency chromosomes for the presence of 2L TAS by in situ hybridization (Table 2). Eight of these showed no evidence for the presence of 2L TAS, and 10 others showed only weak hybridization to the 2L TAS probe (Figure 2)
Identification of 2L TAS on deficiency chromosomes. Arrows point to TAS on the balancer chromosomes. Arrowheads point to the site where TAS should be on the deficiency chromosomes. (A) Df 20/Gla. (B) Df 157/SM1. Deficiency designation is as in Table 1.
. Thus, a substantial proportion of the suppressor chromosomes lack much or all of the 2L TAS array and are considered to be false positives (Table 1, FP4). This proportion could be even higher than these data indicate, because a partial deficiency for the TAS repeat sufficient to cause suppression of TPE may not be obvious from in situ hybridization. Consistent with this idea, three chromosomes, Df 60, 144, and 177, with suppressors that map to the tip of 2L, suggesting a disruption of 2L TAS (Golubovskyet al. 2001; Masonet al. 2003b), show relatively strong hybridization to TAS. Some of the deficiencies in Table 2 could not be tested for 2L TAS, because they did not produce satisfactory salivary chromosome spreads.
Lethal complementation:
Many of the identified deficiencies for 2L TAS are also mutant for the adjacent gene, l(2)gl (Golubovskyet al. 2001). We, therefore, crossed the chosen deficiencies to l(2)gl mutants to inquire into the ability of the deficiency chromosomes to complement the lethality associated with this locus. Two alleles of l(2)gl in different genetic backgrounds were chosen to obviate genetic background effects. The deficiency chromosome must be lethal in combination with both alleles to show a failure to complement. Most of the deficiencies tested complemented both alleles of l(2)gl. Five deficiencies did not complement either allele. Two deficiencies (Df 8 and Df 110) failed to complement the l(2)gl mutations and also failed to hybridize the 2L TAS probe. Df 153 failed to complement the l(2)gl mutations, carries a suppressor that mapped to the 2L tip, but was not tested for the presence of 2L TAS. Interestingly, Df 60 fails to complement the l(2)gl mutations and carries a suppressor that mapped to the tip of 2L, but it also hybridizes to 2L TAS. Given the evidence that a partial or complete deletion of 2L TAS acts as a suppressor of TPE silencing, these latter results suggest that the deletion need not show a discernible decrease in 2L TAS hybridization to exhibit a suppressor phenotype. Finally, Df 81 fails to complement the l(2)gl mutations, but could not be tested in the other assays. Given the results on the other deficiencies, we believe that this gives a strong reason to doubt that the suppressor on this chromosome is a result of the deficiency itself, and this chromosome should be considered a false positive (FP5) until demonstrated otherwise. Failure to complement l(2)gl mutations and to hybridize strongly to the 2L TAS probe in situ suggests a suppressor at the tip of 2L, but does not unequivocally demonstrate that a deficiency does not uncover a suppressor. Thus, these deficiencies (FP4 and FP5) were dropped from further consideration, rather than used to identify regions devoid of suppressors.
Given the high frequency of false positive results, we feel uncomfortable assigning sites of potential TPE suppressors on the basis of equivocal results. We, therefore, chose to ignore deficiencies associated with a suppressor phenotype that overlaps wild type (Table 1, FP6).
A map of potential suppressors of telomeric silencing:
In several regions with potential suppressor deficiencies the ambiguity surrounding the deficiency breakpoints of nonsuppressing deficiencies raised the possibility that the latter deficiencies might overlap. Overlaps would eliminate the ambiguous regions as potential sites of TPE suppressors. We, therefore, looked for genetic evidence for overlaps. The FlyBaseConsortium (2003) reports that complementation tests between deficiencies and gene mutations indicate that Df 174 and 177 in cytological region 55 overlap, and Df 300 and 301 in region 84 also overlap, thus eliminating two potential sites of a suppressor of TPE. We conducted lethal complementation tests between Df 189 and 193 in region 59 and found that they complement, indicating that these two deficiencies do not overlap. Complementation tests indicate that Df 269 and 270 overlap, eliminating a potential suppressor in region 76.
After eliminating false positive and potential false positive results, a map was constructed to identify chromosomal regions that may contain suppressors of TPE (Figures 3 and 4)
Deficiencies for chromosome 2 and their ability to suppress telomeric silencing. Open rectangles below the polytene chromosome represent deficiencies without the ability to suppress TPE. Solid rectangles show deficiencies on chromosomes that suppress TPE. The minimum number of nonsuppressing deficiencies covering the full genetic distance is shown; i.e., redundant deficiencies are not shown. False positive results are not shown. Symbols shown above the chromosome are genes whose mutations exhibit effects on gene expression or genes that are homologous of yeast genes that encode telomeric proteins. Numbers above the chromosome indicate sites identified by the deficiencies as potential sites of suppressors of TPE.
Deficiencies for chromosome 3 and their ability to suppress telomeric silencing. Symbols are the same as for Figure 3.
. Twelve sites of potential TPE suppressors were identified on chromosome 2, and 6 on chromosome 3. Given the high frequency of false positive results and the inability to test all of the deficiency chromosomes adequately, these are probably high estimates for the actual number of suppressors on these chromosomes. As deficiencies for 2L TAS are strong TPE suppressors (Golubovskyet al. 2001; A. Y. Konev and J. M. Mason, unpublished results), and deficiencies for other autosomal telomeres may not exhibit the same phenotype (M. D. Golubovsky, S. Prasad and J. M. Mason, unpublished results), the estimate for the number of suppressor sites on chromosome 2 may be especially high.
To ask whether potentially interesting genes might lie in the regions identified, we placed selected categories of genes on the same map. These include suppressors and enhancers of PEV, PcG, and trithorax group (trxG) genes; genes necessary for RNAi; homologs of genes that encode yeast telomeric proteins; genes that encode components of the nuclear lamin and nuclear pores; and genes that encode post-translational histone modifiers. Of 108 autosomal genes examined, 2 fell into potentially interesting sites identified by the deficiencies. These are Psc on chromosome 2, and gpp, the homolog of yeast DOT1, on chromosome 3. The sites of potential TPE suppressors, as defined here by the deficiencies, encompass ∼5% of the autosomal genome. On the basis of random sampling of the tested loci, we would have expected 6 genes to be in potentially interesting sites. We found 2, suggesting that these “hits” may be due to chance.
Several reports (Crydermanet al. 1999; Kurenovaet al. 1998; Boivinet al. 2003) have implicated the deficiency Su(z)25 as a suppressor of TPE. We, therefore, tested it for suppression and mapped the suppressors and lethals on this chromosome. A strong suppressor was found to be inseparable from al. A second suppressor in the stock could not be mapped easily in the presence of the strong suppressor. This second suppressor segregated independently of al and did not segregate with either the X chromosome or chromosome 3. Thus, it may be on 2R [near Su(z)2] or chromosome 4. There were also multiple lethals on the Su(z)25 chromosome. One mapped to the tip of 2L and failed to complement mutations for l(2)gl. Another lethal appeared to map to the Su(z)2 locus. Most of the tested chromosomes were noncrossover, however, and detailed mapping was not pursued. The Su(z)25 chromosome also failed to hybridize to the 2L TAS. By these assays, it appears that one suppressor is associated with the 2L telomere and not related to the Su(z)25 deficiency, although we cannot exclude the possibility that the second suppressor is a result of the deficiency of the Su(z)2 locus.
A recent report (Boivinet al. 2003) also implicates Psc1, a mutation in another locus uncovered by the Su(z)25 deficiency, as a suppressor of TPE. We found that the suppressor on this chromosome maps to the tip of 2L, not to the Psc locus. This chromosome, however, complements the lethality of l(2)gl mutations.
DISCUSSION
As part of a systematic search for genes in Drosophila that play a role in TPE, we screened the Bloomington autosomal deficiency kits for dominant suppressors of telomeric silencing. Of 372 deficiencies tested, 149 chromosomes gave a positive response. The suppressors on 124 (83%) of the latter are not associated directly with the deficiency itself, but appear to be due to a second mutation on the deficiency chromosome. Ignoring deficiencies on chromosomes with a suppressor phenotype that overlaps wild type, we are left with 25 deficiencies that identify 18 potential sites of TPE suppressors. On chromosome 2, where there were more deficiency chromosomes with a suppressor phenotype and more tools to characterize them, 80% of the suppressors (67/84) were determined to be false positives, while on chromosome 3 more than half (10/18) were false positives. Given the high frequency of false positive results and the inability to adequately test all of the deficiency chromosomes, we may have overestimated the number of suppressor genes.
2L TAS plays a role in TPE:
All nine of the suppressors mapped by meiotic recombination are at or near the 2L telomere. In situ hybridization studies indicated that 18 of 36 deficiency chromosomes tested lacked all or most of the 2L TAS array, independent of the position of the deficiency. Given published observations that even partial deficiencies for 2L TAS may have strong suppressing effects on TPE (Golubovskyet al. 2001), and the present data showing that for half (3/6) of the chromosomes tested in both assays the suppressor that mapped to the 2L telomere region did not show a discernible decrease in TAS hybridization, this proportion (18/36) is likely an underestimate of the number of suppressor chromosomes that lack at least part of the 2L TAS. Taken together, these data reinforce and extend previous suggestions that 2L TAS plays a major role in TPE (Golubovskyet al. 2001; Masonet al. 2003b). It is possible that the suppressive effect of 2L TAS deficiencies on silencing of P{wvar}KR3-2 is the result of homologous interactions. These deficiencies, however, also suppress TPE at nonhomologous telomeres, while the converse is not true; Df of 3R TAS do not suppress TPE at 2L (M. D. Golubovsky, S. Prasad and J. M. Mason, unpublished results). Thus, we believe that deficiencies for 2L TAS have a global impact on telomeric silencing.
2L TAS hybridizes in situ with both the 2L and 3L chromosome tips, but not with the tips of XL, 2R, or 3R (Mechler et al. 1985; Walter et al. 1995), suggesting a similar sequence for the former two TAS arrays. Sequencing of BACs derived from the Drosophila Genome Project also indicates strong similarities between 2L and 3L TAS arrays (A. Villasante, personal communication). We speculate that deficiencies for 3L TAS may have a suppressor phenotype similar to that seen with deficiencies for 2L TAS, and that deficiencies of 3L TAS may be responsible for the high frequency of false positives we find on chromosome 3. The difference in frequency between false positives on chromosomes 2 and 3, that is, presumptive 2L and 3L TAS deficiencies, is consistent with the observation that l(2)gl mutants exist at a high frequency in natural populations (Golubovsky 1978) and that these mutations are primarily terminal (i.e., TAS) deficiencies (Mechleret al. 1985; Walter et al. 1995). It is possible that the terminal 2L region is more susceptible to loss, or that heterozygous deficiencies for the 2L tip region have a selective advantage (Golubovsky 1978).
The role of known genes on TPE:
In an effort to ask whether other suppressors of genetic silencing may act on telomeres, we compared the map positions of Su(var) and PcG genes, as well as their opposites, E(var) and trxG genes, with the loci identified by the deficiencies. RNAi and histone modification may play a role in heterochromatin formation; we therefore considered genes that control these two processes. The position of telomeres in the nucleus, and especially proximity to nuclear pores, is important for telomeric silencing in yeast (Gottaet al. 1996; Galyet al. 2000). We, therefore, examined genes that encode the structural components of the nucleus, including nuclear pores, and lamin, as well as homologs of yeast genes that encode telomere-specific proteins. On the basis of the size of the sites of potential suppressors of TPE and the number of genes considered, and assuming random positions for these genes, we would have expected approximately six genes to lie in these sites; only two (Psc and gpp) fell within these sites. Therefore, the positions of these genes within sites of potential suppressors is not a strong indicator that the genes thus identified are important for telomeric silencing. Indeed, Crydermanet al. (1999) showed that several mutant alleles of Psc do not have suppressor phenotypes. Thus, this gene is probably not involved in telomeric silencing.
Boivinet al. (2003) reported that several PcG and trxG mutations have an effect on telomeric silencing. This observation seems at odds with the present report. How do we interpret the apparent discrepancy? Boivinet al. (2003) tested several mutant alleles for genes examined. In several cases, one or more mutations exhibited a suppressor or enhancer phenotype and other alleles for the gene did not. Standard genetic practice is to assume that phenotypic differences between alleles in different genetic backgrounds are due to background effects until proven otherwise. These authors did the opposite. Their approach seems destined to maximize the number of false positive results. We reexamined two of the three second chromosome mutations they claim suppress telomeric silencing, Su(z)25 and Psc1, and showed that both of these mutant chromosomes have a suppressor at the tip of 2L, even though the mutation being tested was on 2R. Thus, the results of Boivinet al. (2003) require verification. Interestingly, previous results (Crydermanet al. 1999) showed that a Psc1 chromosome did not have a suppressor phenotype. It should be stressed that genetic background effects can be a serious problem when dealing with mutations from different sources.
Numerous genes act to remodel and repress chromatin in heterochromatin and around euchromatic genes during development. Many of these have been identified by dominant mutations that suppress this repression. We have found relatively few potential sites for genes that have a similar effect at telomeres. There are several possible reasons for the difference. First, the 18 sites we have mapped may all identify suppressor genes. This is unlikely, because for nine of nine second chromosomes on which we mapped the suppressor by meiotic recombination, the suppressor was located at the tip of 2L, rather than at the site of the deficiency on that chromosome. Thus, we believe that the remaining deficiencies overestimate the number of suppressor loci. Some of the remaining sites may still contain suppressor genes. The Drosophila homolog of the yeast DOT1 gene is a candidate identified by the deficiencies. Several newly induced mutant alleles for this gene, renamed grappa (gpp), also have a suppressor phenotype (G. Shanower and P. Schedl, personal communication).
Second, we looked at the major autosomes, but the suppressors of telomeric silencing may be on the X or fourth chromosomes. R. Levis (personal communication), for example, has found that several mutant alleles of the X-linked gene ph exhibit a suppressor phenotype and that this phenotype is rescued by a duplication for the region. Although there may be some suppressors on the X, the major autosomes make up 80% of the genome and would be expected to carry a majority of suppressor genes, if they are randomly distributed. Third, autosomal suppressor genes may exist in regions that have not been uncovered by the deficiencies. Although this is possible, the deficiencies we examined span 77% of the two major autosomes. Thus, we should have found a majority of suppressor loci, if they are distributed randomly throughout the genome.
Fourth, partial elimination of the relevant proteins may not suppress TPE. It is possible that suppressors of TPE are recessive, even though many Su(var) and PcG mutations are dominant (Reuter and Wolff 1981; Sinclairet al. 1989; Reuter and Spierer 1992; Pirrotta 1995), and gpp mutations are dominant suppressors of TPE (G. Shanower and P. Schedl, personal communication).
Fifth, we may have chosen the wrong phenotype. As we do not know the mechanism of silencing, it is possible that many mutations in the process decrease, rather than increase, gene expression in telomeric regions. While we did not score for enhancers specifically, they would have been visible in our screen. Other, more subtle phenotypes are also possible.
Finally, chromatin structure in telomeric regions of Drosophila, at least in and around the TAS array, may be simple, with relatively few components. Further searches for disruption of telomeric silencing may reveal that there are, in fact, few genes that play a role in TPE.
TPE is independent of the chromosome capping complex:
A few components of the telomere capping complex have recently been identified. Heterochromatin protein 1 (HP1) binds to chromosome ends in Drosophila independently of the presence of the terminal transposon array or the TAS repeats (Fantiet al. 1998; Siriacoet al. 2002). Null mutations in Su(var)205, the gene that encodes HP1, cause an increase in the length of the terminal HeT-A/TART array when heterozygous (Savitskyet al. 2002) and telomere fusions when homozygous (Fantiet al. 1998). HP1 associates with HP1 origin recognition complex-associated protein (HOAP; Shareefet al. 2001) at chromosome ends (Baduguet al. 2003; Cenciet al. 2003). Disruption of caravaggio (cav), the gene that encodes HOAP, has an effect at telomeres similar to Su(var)205 mutations (Cenciet al. 2003; P. Georgiev, personal communication). Mutations for tefu, the Drosophila ATM homolog, interfere with the binding of HP1 and HOAP to chromosome ends, and mutants for this gene have a phenotype similar to Su(var)205 and cav mutants. It has been proposed that these genes encode components of the telomere capping complex. Mutation or deletion of none of these genes, however, affects TPE dominantly (Crydermanet al. 1999 and this report). Thus, the terminal capping protein complex may act independently of any chromatin complex that plays a role in TPE.
Footnotes
Communicating editor: M. J. Simmons
Acknowledgement
We thank Larry Champion and Jane Koo for excellent technical assistance and Pavel Georgiev and Greg Shanower for sharing unpublished information. We also thank Harald Biessmann and Kevin Cook for comments on the manuscript and Lori Wallrath and Stepháne Ronsseray for their generous gifts of stocks.
Note added in proof: Oikemus et al. (S. R. Oikemus, N. McGinnis, J. Queiroz-Machado, H. Tukachinsky, S. Takada et al., 2004, Drosophila atm/telomere fusion is required for telomeric localization of HP1 and telomere position effect. Genes Dev. 18: 1850–1861) report that mutations in tefu, the Drosophila ATM homologue, reduce the amount of HP1 at telomeres and cause a recessive suppression of telomeric silencing. This suggests an interaction between the telomere capping complex and telomeric silencing.
References
Badugu, R., M. M. Shareef and R. Kellum,
Baur, J. A., Y. Zou, J. W. Shay and W. E. Wright,
Blackburn, E. H.,
Boivin, A., C. Gally, S. Netter, D. Anxolabehere and S. Ronsseray,
Boulton, S. J., and S. P. Jackson,
Cenci, G., G. Siriaco, G. D. Raffa, R. Kellum and M. Gatti,
Cryderman, D. E., E. J. Morris, H. Biessmann, S. C. R. Elgin and L. L. Wallrath,
Dubrana, K., S. Perrod and S. M. Gasser,
Fanti, L., G. Giovinazzo, M. Berloco and S. Pimpinelli,
FlyBase Consortium,
Galy, V., J. C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou et al.,
Gehring, W. J., R. Klemenz, U. Weber and U. Kloter,
Golubovsky, M. D.,
Golubovsky, M. D., A. Y. Konev, M. F. Walter, H. Biessmann and J. M. Mason,
Gotta, M., T. Laroche, A. Formenton, L. Maillet, H. Scherthan et al.,
Gottschling, D. E., O. M. Aparicio, B. L. Billington and V. A. Zakian,
Hazelrigg, T., R. W. Levis and G. M. Rubin,
Horn, D., and G. A. M. Cross,
Karpen, G. H., and A. C. Spradling,
Kurenova, E., L. Champion, H. Biessmann and J. M. Mason,
Kyrion, G., K. Liu, C. Liu and A. J. Lustig,
Laroche, T., S. G. Martin, M. Gotta, H. C. Gorham, F. E. Pryde et al.,
Levis, R., T. Hazelrigg and G. M. Rubin,
Levis, R. W., R. Ganesan, K. Houtchens, L. A. Tolar and F. M. Sheen,
Lindsley, D. L., and G. G. Zimm, 1992 The Genome of Drosophila melanogaster. Academic Press, San Diego.
Marin, L., M. Lehmann, D. Nouaud, I. Hassan, D. Anxolabéhère et al.,
Mason, J. M., and H. Biessmann,
Mason, J. M., A. Haoudi, A. Y. Konev, E. Kurenova, M. F. Walter et al.,
Mason, J. M., A. Y. Konev and H. Biessmann,
Mason, J. M., A. Y. Konev, M. D. Golubovsky and H. Biessmann,
Mechler, B. M., W. McGinnis and W. J. Gehring,
Moretti, P., K. Freeman, L. Coodly and D. Shore,
Nimmo, E. R., G. Cranston and R. C. Allshire,
Palladino, F., and S. M. Gasser,
Park, M. J., Y. K. Jang, E. S. Choi, H. S. Kim and S. D. Park,
Pirrotta, V.,
Pryde, F. E., H. C. Gorham and E. J. Louis,
Reuter, G., and P. Spierer,
Reuter, G., and I. Wolff,
Roseman, R. R., E. A. Johnson, C. K. Rodesch, M. Bjerke, R. N. Nagoshi et al.,
Rudenko, G., P. A. Blundell, A. Dirksmulder, R. Kieft and P. Borst,
Savitsky, M., O. Kravchuk, L. Melnikova and P. Georgiev,
Shareef, M. M., C. King, M. Damaj, R. Badagu, D. W. Huang et al.,
Shore, D.,
Sinclair, D. A., V. K. Lloyd and T. A. Grigliatti,
Siriaco, G. M., G. Cenci, A. Haoudi, L. E. Champion, C. Zhou et al.,
Talbert, P. B., C. D. S. Leciel and S. Henikoff,
Tower, J., G. H. Karpen, N. Craig and A. C. Spradling,
Wallrath, L. L., and S. C. R. Elgin,
Walter, M. F., C. Jang, B. Kasravi, J. Donath, B. M. Mechler et al.,
Weiler, K. S., and B. T. Wakimoto,
Author notes
Present address: Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX 75390.
Present address: Postgenomics, San Diego, CA 92121.