-
PDF
- Split View
-
Views
-
Cite
Cite
N Carol Casavant, LuAnn Scott, Michael A Cantrell, Lara E Wiggins, Robert J Baker, Holly A Wichman, The End of the LINE?: Lack of Recent L1 Activity in a Group of South American Rodents, Genetics, Volume 154, Issue 4, 1 April 2000, Pages 1809–1817, https://doi.org/10.1093/genetics/154.4.1809
Close - Share Icon Share
Abstract
L1s (LINE-1: Long Interspersed Nuclear Element 1) are present in all mammals examined to date. They occur in both placental mammals and marsupials and thus are thought to have been present in the genome prior to the mammalian radiation. This unusual conservation of a transposable element family for over 100 million years has led to speculation that these elements provide an advantage to the genomes they inhabit. We have recently identified a group of South American rodents, including rice rats (Oryzomys), in which L1s appear to be quiescent or extinct. Several observations support this conclusion. First, genomic Southern blot analysis fails to reveal genus-specific bands in Oryzomys. Second, we were unable to find recently inserted elements. Procedures to enrich for young elements did not yield any with an intact open reading frame for reverse transcriptase; all elements isolated had numerous insertions, deletions, and stop codons. Phylogenetic analysis failed to yield species-specific clusters among the L1 elements isolated, and all Oryzomys sequences had numerous private mutations. Finally, in situ hybridization of L1 to Oryzomys chromosomes failed to reveal the characteristic L1 distribution in Oryzomys with either a homologous or heterologous probe. Thus, Oryzomys is a viable candidate for L1 extinction from a mammalian host.
RETROTRANSPOSONS related to L1 (LINE-1: Long Interspersed Nuclear Element 1) are found across a wide phylogenetic range—in protists, fungi, plants, insects, fish, amphibians, and mammals (Xiong and Eickbush 1990; Duvernell and Turner 1998). They make up a significant proportion of the mammalian genome, estimated at tens of thousands to a hundred thousand copies in species for which careful studies have been made (Hutchison III et al. 1989). In rats, it is proposed that in the past 10 million years alone, rat genome size has increased 10–20% due to the insertion of L1 elements (Pascale et al. 1990). In situ hybridization shows that the elements are nonuniformly distributed on all chromosomes, with heavy accumulation in the A + T-rich G-bands and sometimes in the sex chromosomes (Korenberg and Rykowski 1988; Wichman et al. 1992).
L1s are present in all mammals examined to date and are thought to have been present in the genome prior to the mammalian radiation (Burton et al. 1986). All evidence indicates that L1 elements are transmitted vertically, from parent to offspring, rather than horizontally by infectious transmission. Therefore, active copies must be maintained in the genome if L1 is to persist over evolutionary time. In humans and Mus, most insertions are 5′ truncated, and many others have debilitating mutations that render them inactive. Only a minority of the thousands of elements in the genome is capable of producing copies. As an active lineage accumulates new mutations, this variation is passed on to all subsequent members of the lineage and can serve as a taxonomic character to study the evolution of both the element and its host (Pascale et al. 1993; Casavant and Hardies 1994; Furano and Usdin 1995; Casavant et al. 1996; Modi 1996). Analysis of L1s from a number of species suggests that newly inserted elements belong to one or a few closely related lineages (Martin et al. 1985; Rikke et al. 1991; Casavant and Hardies 1994; Casavant et al. 1996; Cabot et al. 1997; Naas et al. 1998). It is paradoxical that L1s maintain a small number of active lineages without undergoing occasional extinction from their mammalian host.
Most aspects of L1 biology appear to fit the selfish gene model. Insertions of elements into the genome cause mutations in host genes, provide sites for nonhomologous recombination, and are thought to provide reverse transcriptase for the movement of pseudogenes and other retroelements. Thus, it appears that L1 elements are maintained at a cost to the host. However, the fact that these elements are ubiquitous among those genomes studied has been interpreted to suggest that they may have a function in the host genome. Potential roles include the generation of genetic diversity and a role in the repair of chromosomal breaks (Hutchison III et al. 1989).
Although no group of mammals has been documented to lack L1 activity, a dramatic decrease in activity (or extinction of L1) has been proposed in voles (Vanlerberghe et al. 1993). This conclusion was based on sequence analysis of randomly selected L1 elements. However, genomic Southern blot analysis (Modi 1996) revealed a large number of species-specific bands in voles, suggesting a high level of recent L1 activity. Subsequent sequence analysis of vole L1s, using methods that enrich for younger insertions (R. Grahn and H. A. Wichman, unpublished results), confirms these results. Thus, conclusions about L1 activity are particularly sensitive to sampling methods.
While it is always easier to demonstrate the recent activity of a family of transposable elements than to document its extinction, any viable candidate for L1 extinction should meet the following criteria:
Genomic Southern blot analysis fails to reveal taxon-specific bands. The number and rank of taxa with an absence of novel bands will demarcate the extinction event.
Recently inserted elements cannot be isolated by protocols designed to enrich for young elements. Recently inserted elements can be recognized by the following: (i) an intact open reading frame (ORF), (ii) conservation of amino acid sequence, (iii) monophyletic species cluster within an L1 lineage, and (iv) few private mutations as deduced by phylogenetic analysis.
In situ hybridization fails to reveal the characteristic, genome-wide pattern of L1 distribution.
Using genomic Southern blot analysis, we have serendipitously identified a group of South American rodents, including the rice rat (Oryzomys) and the marsh rat (Holochilus), that appears to have a greatly reduced copy number of L1 elements. In this article, we apply the above criteria to explore the possibility that these rodents provide a potential model for L1 extinction.
METHODS AND MATERIALS
Specimens examined: Tissues for this study were provided by the Museum, Texas Tech University (rice rat, Oryzomys macconnelli Tk11324 and O. palustris Tk28621; pygmy mouse, Baiomys taylori Tk32211; harvest mouse, Reithrodontomys fulvescens Tk21614; deer mouse, Peromyscus maniculatus Tk27553), Museum of Natural Sciences, Louisiana State University (wood rat, Neotoma floridana LSUMZ27523; rice rat, O. palustris LSUMZ28685), Museum of Zoology, University of Michigan (marsh rat, Holochilus brasiliensis UMMZ398), and Savannah River Ecology Laboratory (cotton rat, Sigmodon hispidus). DNA was extracted as previously reported (Casavant et al. 1996).
Genomic Southern blot analysis: Southern blot analysis was carried out as previously described (Casavant et al. 1996). DNA was digested separately with EcoRV, BglII, HaeIII, RsaI, EcoRI, or HincII. Southern blots were probed with L1GEN from Peromyscus (Casavant et al. 1998) or one of the Oryzomys L1 clones described below and scored for the presence of distinct bands visible above the background smear of hybridization.
Isolation of L1 sequences from Oryzomys and Sigmodon: Oryzomys L1 sequences were isolated from a band excised from an agarose gel as previously described (Casavant et al. 1996), except that a 1.8-kb EcoRI band was used because it showed the strongest hybridization to an L1 probe in a genomic Southern blot. L1 sequences were also isolated by PCR as previously described (Casavant et al. 1996). Primers were to conserved regions of L1 reverse transcriptase based on sequences from Peromyscus, the most closely related rodents for which L1s had been characterized at that time, and PCR was carried out under conditions of reduced stringency to allow some primer mismatch. Total genomic DNA or the 1.8-kb EcoRI fragment was used as template.
When these methods failed to yield young L1 elements from Oryzomys, a protocol was developed to enrich for intact ORFs (M. A. Cantrell and H. A. Wichman, unpublished results). PCR primers (Casavant et al. 1996) and the cloning vector pBSIIKS+ (Stratagene, La Jolla, CA) were modified so that when the PCR product was cloned into lacZ using restriction sites included in the primers, it was in the proper orientation and the reverse transcriptase fragment was in the same reading frame as lacZ. Blue-white screening could be used to enrich for clones containing recently inserted elements. Clones maintaining the reading frame tended to yield blue colonies when plated on Xgal and IPTG, while clones with disrupted reading frames tended to yield white colonies. The upstream primer (5′ AAGAATTCCGCAGGATACAAGATCAACTCA 3′) is homologous to bases 5060 through 5080 of GenBank accession no. M13002 (Mus L1) and includes an EcoRI restriction site. The downstream primer (5′ AAGGATCCCAATTCGATTCCATTGGT 3′) corresponds to 5565 through 5582 and includes a BamHI restriction site. Clones from both blue and white colonies were selected and sequenced. These primers were also used to amplify and clone elements from Sigmodon. Sequence was determined on both strands on an ABI 377 automated DNA sequencer.
An alternative set of primers was designed later to incorporate degeneracy at variable bases and amplify a slightly larger region. These primers also incorporated an additional cloning site to accommodate the possible evolution of restriction sites within the region of interest. Design of these modified primers was based primarily on sequences from primates and rodents, but we also made use of L1 sequences from dog and rabbit. The upstream primer (5′ CCACGAATTCCTCGAGCYTITWYGCIGAYGAYATGA 3′) is homologous to bases 4969 through 4987 of GenBank accession no. M13002 (Mus L1) and includes EcoRI and XhoI restriction sites. The downstream primer (5′ GTTGGATCCCTGCAGTGTCIRTICKRTTVYAYTGRTC) corresponds to 5564 through 5583 and includes PstI and BamHI restriction sites.
Sequence analysis: Sequences were aligned using MegAlign 3.1.7 (DNAStar, Madison, WI); alignments were modified by hand. The region examined here corresponds to positions 5093 through 5557 of L1 from Mus (GenBank accession no. M13002). Additional L1 sequences were included in the alignment. For phylogenetic analysis, sequences from house mouse (Mus, GenBank accession no. M13002) and deer mouse (Peromyscus, U43362, U70924, U70929–30, U79033–35) were included. A second alignment was constructed to determine which amino acids are phylogenetically conserved among mammals within the region of reverse transcriptase examined. Only sequences with intact ORFs were included in this alignment. Where multiple such sequences were available, the youngest elements within an L1 clade, as determined by phylogenetic analysis, were used. Sequences from several rodents, primates, bat, and zebra were included, and multiple sequences were used from each species (data not shown). Amino acids were scored as conserved if they were invariant, or if only a single private change appeared among all of the sequences examined. Of the 155 amino acid sites in the region examined, 64 were scored as conserved. These sites were examined in the Oryzomys and Sigmodon L1 sequences.
Phylogenetic trees were constructed by parsimony analysis with PAUP*, version 4.0.0d64 written by David L. Swofford, using a heuristic algorithm with 1000 bootstrap replicates. L1 sequence from Mus was designated as the outgroup.
In situ hybridization: Karyotypes were prepared from bone marrow as previously described (Baker and Qumsiyeh 1988). L1 clones for both Oryzomys and Sigmodon (OryMe41 and Sig8b) were used as probes for in situ hybridization. Probes were labeled by standard nick translation with biotinylated dATP using the GIBCO BRL (Gaithersburg, MD) BioNick nick translation kit following the manufacturer's instructions. Hybridization followed the procedures as previously described (Baker and Wichman 1990). Hybridization patterns were characterized by examining at least 10 complete spreads from each individual using an Olympus (Melville, NY) epi-fluorescent microscope with a dual-band pass filter allowing the simultaneous visualization of propidium iodide and FITC. Photomicrographs were taken using Kodak Royal Gold 1000 color print film. To confirm that differences in hybridization intensity were robust, additional hybridizations were carried out in which cells from the two species were mixed and hybridizations carried out on the same slide.
RESULTS
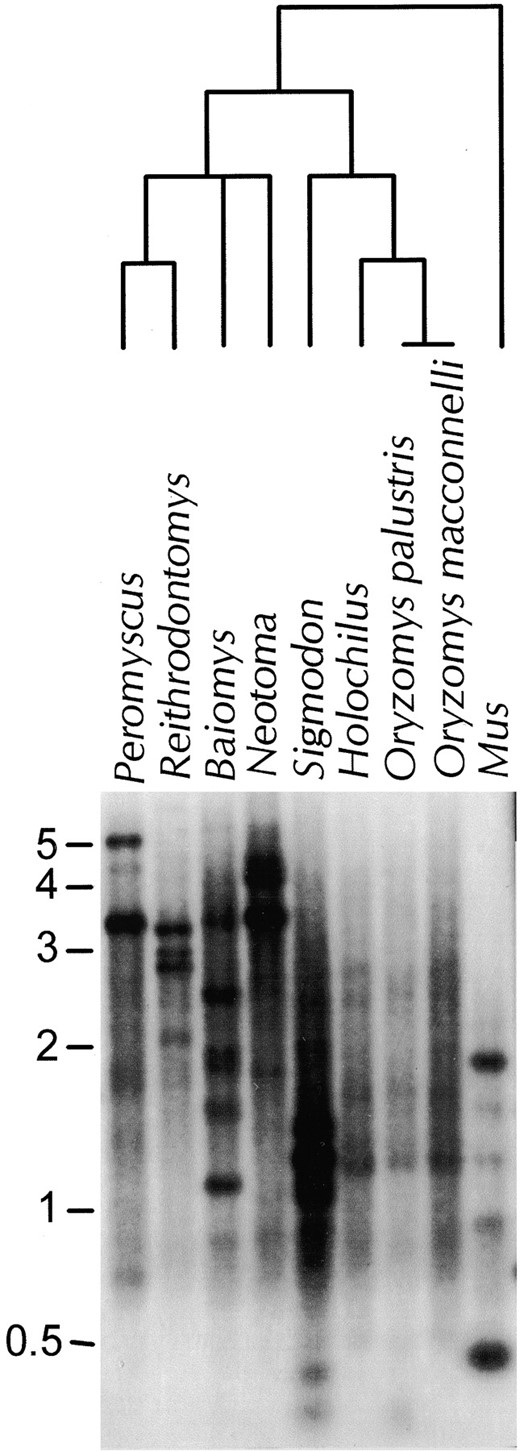
Genomic Southern blot analysis failed to reveal taxon-specific bands: When Southern blot analysis is carried out on mammalian genomic DNA using L1 as a probe, discrete bands are detectable above a background smear of hybridization on the autoradiogram. The presence of such bands indicates that a subset of L1-hybridizing sequences in the genome share two or more restriction sites. Restriction site changes in active elements can give rise in taxon-specific bands, while elements that were inserted prior to species or genus divergence, or were derived from active elements in which no restriction site evolution had occurred, give rise to bands shared across taxa. Taxon-specific bands are a landmark of L1 activity. Genomic Southern blot analysis suggests a paucity of L1 elements in Oryzomys and Holochilus compared to other rodents examined (Figure 1). The detectable bands (and to some extent the background smear) were fainter in Oryzomys and Holochilus than in other species examined, whether the probe was a homologous (Oryzomys) or heterologous (Peromyscus) L1 fragment. Genus-specific bands were detected in Mus, Sigmodon, Neotoma, Baiomys, Reithodontomys, and Peromyscus. These were especially obvious when DNA was digested with enzymes such as HaeIII or RsaI that have 4-bp recognition sites. Based on the sequence of
Distribution of L1 in six genera of rodents. The relationship between genera is shown in the phylogenetic tree above the Southern blot (Carleton 1980; Patton et al. 1981; Smith and Patton 1999). Genomic DNA was digested with HaeIII and probed with L1GEN, a 252-bp subclone of the Man109 L1 clone from Peromyscus maniculatus (Casavant et al. 1998). Species examined include P. maniculatus, Reithrodontomys fulvescens, Baiomys taylori, Neotoma floridana, Sigmodon hispidus, Holochilus brasiliensis, Oryzomys palustris, O. macconnelli, and Mus musculus.
MMU15647 (Martin 1995), the active L1 subfamily in Mus should yield a 1.9-kb band in a HaeIII digestion when probed with L1GEN or any of the clones generated for this study. This band was strongly detected (Figure 1), despite the fact that these heterologous probes are from species much more distant from Mus than from Oryzomys or Holochilus. However, few bands were detected in Oryzomys or Holochilus, and those that were visible were faint, common to both genera, and also detected in Sigmodon and sometimes other, more distantly related genera. Taxon-specific bands were not detected in either Oryzomys or Holochilus with any of the six enzymes used. The conservation of these bands across genera, the faintness of hybridization relative to Sigmodon, and the ability to detect active subfamilies in a much more distant genus suggest that the L1 elements in Oryzomys and Holochilus have not undergone a recent burst of transposition.
Ancient L1 elements were cloned from Oryzomys, but we were unable to isolate young elements: The L1 region analyzed from the Oryzomys genome corresponds to positions 5093 through 5557 of L1 from Mus (GenBank accession no. M13002) and is part of the reverse transcriptase gene. Multiple L1 clones were isolated from O. palustris (designated OryP) and O. macconnelli (designated OryM). Initially, the 1.8-kb EcoRI band, which was distinct and highly conserved across taxa (data not shown), was excised from an agarose gel, DNA was extracted and cloned, and clones were screened with an L1 probe from the deer mouse, Peromyscus. Sequences OryP5, OryP6, OryP40, and OryP41 were acquired by this procedure. Additional sequences were obtained by using primers designed to highly conserved regions of the second ORF of L1 (Casavant et al. 1996). PCR was carried out under conditions of reduced stringency to allow some primer mismatch. Total genomic DNA (for clones OryP3 and OryP4) or the 1.8-kb EcoRI fragment (for OryMC4 and OryME41) was used as template. When these methods failed to yield young L1 elements from Oryzomys, a protocol was developed to enrich for intact ORFs. PCR product was cloned in frame into lacZ, such that clones maintaining the reading frame tended to yield blue colonies when plated on Xgal and IPTG, while clones with disrupted reading frames tended to yield white colonies. Clones from both blue and white colonies were selected and sequenced. For Oryzomys, all six blue colonies selected gave rise to identical clones that were truncated relative to the amplified L1 sequence. White colonies yielded Ory-Ma8w, OryMa20w, one truncated clone, and two sequences identical to OryMa8w. Sigmodon sequences Sig7b, Sig8b, Sig9b, Sig10b, Sig11b, and Sig12b were acquired from blue colonies using genomic DNA from S. hispidus as a PCR template and the lacZ vector system to enrich for intact ORFs. Sig5w was acquired from a white colony for confirmation of the screening technique.
Isolating a region of L1 reverse transcriptase from Oryzomys by any of these methods was considerably more difficult than for other rodent taxa we have examined. To date we have isolated this region of L1 from over a dozen genera of rodents, including Mus, Peromyscus, Sigmodon, Microtus, Phodopus, and others. Despite considerable effort, no Oryzomys clones were found to contain intact ORFs. The data from the lacZ vector is most illuminating. Typically, the use of this vector with other rodent DNAs yields a high percentage of blue colonies (e.g., 64% in Peromyscus and 77% in Sigmodon), and over 80% of these blue clones have intact ORFs. For a variety of technical reasons, these numbers cannot be extrapolated to estimate the percentage of L1 elements in the genome that are active, or even relatively young. For example, PCR itself enriches for younger elements. However, these numbers do indicate that L1 elements have been recently quite active in these rodent genomes. In contrast, only 8% of the clones were blue in Oryzomys, and none yielded intact ORFs. In fact, all six of the clones sequenced from blue colonies were an identical, truncated fragment of 127 bp. This short region had one stop codon, changes at 5 of the 18 conserved amino acid residues within the region, and was 20 to 50% divergent from the remaining Oryzomys L1 isolates. In contrast, all six of the Sigmodon clones sequenced from blue colonies were unique and had intact ORFs.
Ten Oryzomys and seven Sigmodon L1 reverse transcriptase clones, sequenced on both strands, were analyzed in detail. Sigmodon is included because it is the genus most closely related to Oryzomys in which we have demonstrated recent L1 activity by all of the criteria above. Identical clones, one recombinant Oryzomys clone, and the truncated clones discussed above were not included, but all appear to be as ancient as those used in the phylogenetic analysis. We did not attempt to determine if the recombinant clone was a true genomic recombinant or a PCR artifact. The proportion of identical L1 clones obtained by PCR is higher in Oryzomys than in other rodents we have analyzed and may indicate a scarcity of amplifiable templates in the Oryzomys genome.
Alignment of the Oryzomys and Sigmodon L1 sequences revealed that the Oryzomys sequences had from one to eight deletions relative to an intact ORF, 80% of the sequences had one or two insertions, and 90% of the sequences had two or more stop codons within the region examined (Table 1). None of the Sigmodon sequences from blue colonies had deletions, insertions, or stop codons in the region examined, but Sig5w, isolated from a white colony, had one insertion and one stop codon. In addition, the Oryzomys sequences had an average of 11.5 changes (range 6–20) at amino acid residues that were conserved among the mammalian L1 sequences. Sigmodon sequences had an average of 1.7 such changes (range 0–4). Taken together, these data indicate that the Oryzomys L1 sequences we were able to isolate represent ancient fossil inserts, while at least some of the Sigmodon sequences are from recently inserted elements.
Phylogenetic analysis was performed on the Oryzomys,
Analysis of changes in L1 sequences from Oryzomys and Sigmodon
| . | Deletions . | Insertions . | Stop codons . | Changes at conserved sites . | Private mutations . |
|---|---|---|---|---|---|
| OryP3 | 1 | 1 | 2 | 6 | 23 |
| OryP4 | 4 | 2 | 3 | 16 | 52 |
| OryP5 | 6 | 0 | 2 | 15 | 59 |
| OryP6 | 3 | 1 | 0 | 11 | 26 |
| OryP40 | 2 | 2 | 4 | 6 | 26 |
| OryP41 | 2 | 2 | 2 | 11 | 33 |
| OryMa8w | 3 | 1 | 6 | 9 | 39 |
| OryMa20w | 8 | 0 | 4 | 20 | 75 |
| OryMc4 | 5 | 1 | 4 | 13 | 35 |
| OryMe41 | 4 | 1 | 2 | 8 | 32 |
| Sig5w | 0 | 1 | 1 | 6 | 16 |
| Sig7b | 0 | 0 | 0 | 1 | 11 |
| Sig8b | 0 | 0 | 0 | 3 | 8 |
| Sig9b | 0 | 0 | 0 | 1 | 8 |
| Sig10b | 0 | 0 | 0 | 4 | 8 |
| Sig11b | 0 | 0 | 0 | 0 | 4 |
| Sig12b | 0 | 0 | 0 | 1 | 8 |
| . | Deletions . | Insertions . | Stop codons . | Changes at conserved sites . | Private mutations . |
|---|---|---|---|---|---|
| OryP3 | 1 | 1 | 2 | 6 | 23 |
| OryP4 | 4 | 2 | 3 | 16 | 52 |
| OryP5 | 6 | 0 | 2 | 15 | 59 |
| OryP6 | 3 | 1 | 0 | 11 | 26 |
| OryP40 | 2 | 2 | 4 | 6 | 26 |
| OryP41 | 2 | 2 | 2 | 11 | 33 |
| OryMa8w | 3 | 1 | 6 | 9 | 39 |
| OryMa20w | 8 | 0 | 4 | 20 | 75 |
| OryMc4 | 5 | 1 | 4 | 13 | 35 |
| OryMe41 | 4 | 1 | 2 | 8 | 32 |
| Sig5w | 0 | 1 | 1 | 6 | 16 |
| Sig7b | 0 | 0 | 0 | 1 | 11 |
| Sig8b | 0 | 0 | 0 | 3 | 8 |
| Sig9b | 0 | 0 | 0 | 1 | 8 |
| Sig10b | 0 | 0 | 0 | 4 | 8 |
| Sig11b | 0 | 0 | 0 | 0 | 4 |
| Sig12b | 0 | 0 | 0 | 1 | 8 |
Each sequence was examined to determine the number of deletions, insertions, stop codons, changes at conserved amino acid residues, and mutations that map to the terminal branches of the phylogenetic tree (private mutations).
Analysis of changes in L1 sequences from Oryzomys and Sigmodon
| . | Deletions . | Insertions . | Stop codons . | Changes at conserved sites . | Private mutations . |
|---|---|---|---|---|---|
| OryP3 | 1 | 1 | 2 | 6 | 23 |
| OryP4 | 4 | 2 | 3 | 16 | 52 |
| OryP5 | 6 | 0 | 2 | 15 | 59 |
| OryP6 | 3 | 1 | 0 | 11 | 26 |
| OryP40 | 2 | 2 | 4 | 6 | 26 |
| OryP41 | 2 | 2 | 2 | 11 | 33 |
| OryMa8w | 3 | 1 | 6 | 9 | 39 |
| OryMa20w | 8 | 0 | 4 | 20 | 75 |
| OryMc4 | 5 | 1 | 4 | 13 | 35 |
| OryMe41 | 4 | 1 | 2 | 8 | 32 |
| Sig5w | 0 | 1 | 1 | 6 | 16 |
| Sig7b | 0 | 0 | 0 | 1 | 11 |
| Sig8b | 0 | 0 | 0 | 3 | 8 |
| Sig9b | 0 | 0 | 0 | 1 | 8 |
| Sig10b | 0 | 0 | 0 | 4 | 8 |
| Sig11b | 0 | 0 | 0 | 0 | 4 |
| Sig12b | 0 | 0 | 0 | 1 | 8 |
| . | Deletions . | Insertions . | Stop codons . | Changes at conserved sites . | Private mutations . |
|---|---|---|---|---|---|
| OryP3 | 1 | 1 | 2 | 6 | 23 |
| OryP4 | 4 | 2 | 3 | 16 | 52 |
| OryP5 | 6 | 0 | 2 | 15 | 59 |
| OryP6 | 3 | 1 | 0 | 11 | 26 |
| OryP40 | 2 | 2 | 4 | 6 | 26 |
| OryP41 | 2 | 2 | 2 | 11 | 33 |
| OryMa8w | 3 | 1 | 6 | 9 | 39 |
| OryMa20w | 8 | 0 | 4 | 20 | 75 |
| OryMc4 | 5 | 1 | 4 | 13 | 35 |
| OryMe41 | 4 | 1 | 2 | 8 | 32 |
| Sig5w | 0 | 1 | 1 | 6 | 16 |
| Sig7b | 0 | 0 | 0 | 1 | 11 |
| Sig8b | 0 | 0 | 0 | 3 | 8 |
| Sig9b | 0 | 0 | 0 | 1 | 8 |
| Sig10b | 0 | 0 | 0 | 4 | 8 |
| Sig11b | 0 | 0 | 0 | 0 | 4 |
| Sig12b | 0 | 0 | 0 | 1 | 8 |
Each sequence was examined to determine the number of deletions, insertions, stop codons, changes at conserved amino acid residues, and mutations that map to the terminal branches of the phylogenetic tree (private mutations).
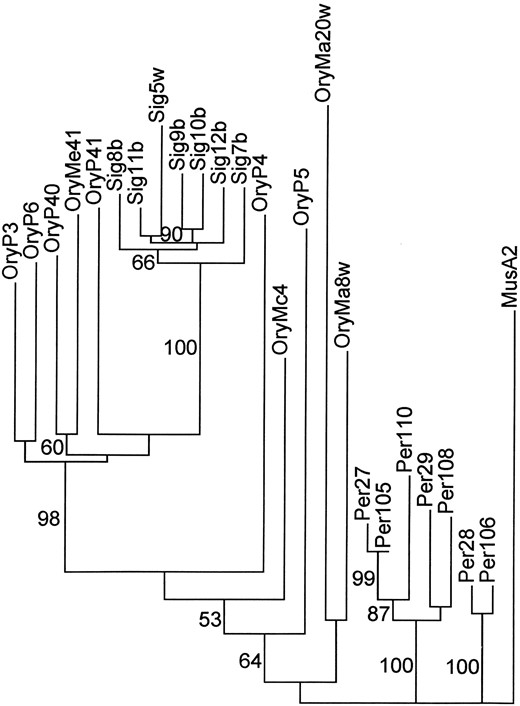
Sigmodon, and Peromyscus L1 sequences, Mus L1 (Casavant et al. 1996), as an outgroup. A phylogram of the 50% majority rule consensus tree is shown in Figure 2. Bootstrap values above 50% are shown. Both of the previously identified Peromyscus lineages (Casavant et al. 1996) formed a monophyletic group with 100% bootstrap support and short terminal branch lengths. Sigmodon sequences also formed a monophyletic clade with 100% support and short terminal branch lengths. Oryzomys sequences clustered with Sigmodon sequences, but some elements appear to have arisen before the divergence between the two genera. The relationship among Oryzomys sequences was not well resolved; few informative characters support relationships among any group of Oryzomys sequences. Such informative characters represent changes in the master elements that gave rise to the copies sampled (Casavant et al. 1996). This lack of informative characters suggests that there has been little evolution of active master elements in Oryzomys since its split from Sigmodon, whereas Sigmodon sequences share 24 changes in the lineage of their master elements in the same time period. Finally, the short terminal branch lengths on the youngest Sigmodon and Peromyscus L1 sequences indicate that they have accumulated few private mutations since inserting into the genome. In contrast, even the youngest Oryzomys sequences have accumulated at least 5% change since inserting into the genome, adding additional support to the view that these insertions are ancient. Isolation of additional clones using the modified degenerate primers failed to yield more recently inserted elements (data not shown).
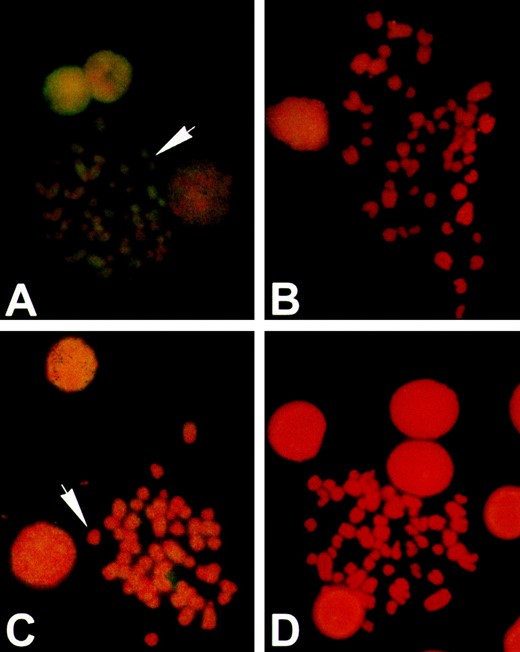
Fluorescent in situ hybridization (FISH) with L1 probes revealed consistent differences in distribution and amount of signal between Sigmodon and Oryzomys chromosomes: The Sigmodon L1 probe provided a hybridization pattern in Sigmodon (Figure 3A) comparable in both intensity and distribution to that reported for L1 elements in other rodent species (Boyle et al. 1990; Wichman et al. 1992; Baker and Kass 1994), although accumulation on the sex chromosomes is not apparent here. Specifically, L1 was highly repetitive on all chromosomes and produces a banding pattern, but hybridization was lacking in heterochromatic regions of the chromosomes. Examples of heterochromatic exclusion are indicated by the white arrows in Figure 3, A and C. The Oryzomys L1 probe on Sigmodon chromosomes (Figure 3C) showed a similar hybridization pattern but a weaker signal. Both Sigmodon (Figure 3B) and Oryzomys (Figure 3D) probes hybridized only faintly to Oryzomys chromosomes. The signal is most easily compared by examining the extent of hybridization to interphase nuclei, visible as discrete circles in A–D. It may seem paradoxical that the signal on the Oryzomys chromosomes was slightly more apparent when a Sigmodon L1 clone was used as a probe (Figure 3B) than when an Oryzomys L1 clone was used (Figure 3D). We believe this reflects the fact that Oryzomys L1s have been accumulating mutations at the neutral rate for a long period, while the youngest Sigmodon L1s have been under selection to maintain their ability to transpose. In particular, the presence of multiple indels in the Oryzomys elements may interfere with hybridization, especially when they are present in both the target
Majority rule tree L1 sequences from Oryzomys and Sigmodon. Tree is rooted with L1 sequences from Mus and Peromyscus. Parsimony analysis was carried out using a heuristic algorithm with 1000 bootstrap replicates; bootstrap values >50% are shown. Branch lengths are proportional to the number of changes that support eachclade. Terminal branch lengths are proportional to the number of private mutations in each element.
and probe sequences as they are when Oryzomys chromosomes are probed with an Oryzomys L1 clone.
DISCUSSION
Most aspects of L1 biology and evolution are consistent with the selfish parasite model. However, some aspects of L1 evolution are enigmatic for a selfish element. In particular, it is difficult to understand how L1s are maintained in the genome as a small number of active lineages (Martin et al. 1985; Rikke et al. 1991; Casavant and Hardies 1994; Casavant et al. 1996; Cabot et al. 1997; Naas et al. 1998), especially in light of the long-term evolutionary persistence of L1 in mammals. The maintenance of few active lineages without eventual L1 extinction requires a fine balance between the rate at which new active templates are produced and the rate at which old templates become inactive. If new active elements are produced more frequently than old ones lose activity, multiple lineages will evolve. If they are produced less frequently, the L1 family will become extinct in that species. It is difficult to imagine how this fine balance is maintained, especially in light of evidence that L1 transposition occurs in bursts (Pascale et al. 1993; Verneau et al. 1998).
In humans, careful estimates of the number of potentially
In situ hybridization of rodent chromosomes with L1 probes. Sigmodon (A and C) and Oryzomys (B and D) chromosomes probed with L1 sequences Sig8b from Sigmodon (A and B) and OryMe41 from Oryzomys (C and D). Chromosomes were stained with propidium iodide (red) and probes were labeled by biotinylation and detected by immunofluorescence (yellow). Hybridization to L1 probes in Sigmodon is typical of the pattern seen in other mammals. Hybridization to Oryzomys is reduced compared to Sigmodon and is most evident in interphase nuclei.
active L1s suggest that between 30 and 60 loci per diploid genome are sequence competent for transposition (Sassaman et al. 1997), and yet these loci vary by only a few percent and are clearly members of a single, closely related lineage. In Mus, the number of potentially active L1 loci per diploid genome has been estimated at 3000, and again these appear to be closely related (DeBerardinis et al. 1998). These estimates are based on an in vitro assay. Genomic position or regulatory mechanisms such as methylation may silence an element that is otherwise transposition competent, so this likely represents an overestimate of the number of loci active in vivo. Nevertheless, this number is larger than one might expect given the close relationship between the youngest insertions into the genome. We have yet to find a mammal in which the active loci have diverged to the extent one would expect for a selfish element, nor have we found a mammal in which L1 sequences are entirely absent.
Maintenance of L1s in the genome as a small number of lineages, but without frequent extinction of the elements from some host genomes, must require some level of regulation by the host, the elements, or both. Methylation (Liu and Schmid 1993; Hata and Sakaki 1997; O'Neill et al. 1998), and, more recently cosuppression (i.e., post-transcriptional silencing of multicopy genes—Bingham 1997; Pal-Bhadra et al. 1997; Matzke and Matzke 1998; Jensen et al. 1999), have been suggested as possible mechanisms for the control of transposable elements. Because cosuppression would be correlated with the level of transcription, suppression might be greatest when there are large numbers of active templates and be reduced when the number of active templates falls. Thus, during periods of cosuppression, few new templates would be produced, but when the number of active elements in a species falls below some critical level, cosuppression would be lifted and many new, closely related templates might be produced. Cosuppression may thus have properties that are especially attractive to explain the enigmatic characteristics of L1 lineages discussed above, but the details of this phenomenon are not well understood and its role in regulating L1 expression remains speculative.
Here we present data to suggest that Oryzomys is the first viable candidate for L1 extinction in mammals. A prior report of L1 slowdown or extinction in voles was based on an analysis of randomly selected elements (Vanlerberghe et al. 1993), but several lines of evidence suggest that L1s are not extinct in voles. First, genomic Southern blot analysis revealed that fully half of the distinct bands were species specific in voles (Modi 1996). This suggests a high level of recent transposition. Second, using methods similar to those described here, we have isolated numerous sequences from Microtus arvalis, which have intact ORFs. The youngest of these sequences form monophyletic species clusters and have few private mutations as deduced by phylogenetic analysis (R. Grahn and H. A. Wichman, unpublished results). Thus, on the basis of the criteria listed above, we were able to rule out Microtus as a viable candidate for L1 extinction. By these same criteria, Oryzomys remains a viable candidate for L1 extinction. First, genomic Southern blot analysis fails to reveal genus-specific bands in Oryzomys or in the closely related genus Holochilus. Furthermore, the apparent copy number of L1s in bands conserved across genera was much lower in these species than in other rodents examined. Second, despite considerable effort using methods that have detected young elements in numerous other species, we were unable to find recently inserted elements in Oryzomys. None of the elements isolated had intact ORFs. All elements examined in a 465-bp region of ORF2 had deletions, and most had insertions and stop codons. Amino acid residues conserved across rodents, primates, bats, and equids were not conserved among the elements examined from Oryzomys. Phylogenetic analysis failed to yield species-specific clusters among the L1 elements isolated, and all Oryzomys sequences had numerous private mutations. Finally, in situ hybridization of L1 failed to reveal the characteristic strong pattern of genome-wide distribution in Oryzomys with either a homologous or heterologous probe.
To approximate the age of the youngest L1 elements isolated from Oryzomys, we estimated the time of insertion by counting the number of private mutations on each terminal branch of the phylogenetic tree. The minimal estimate of private mutations ranged from 23 to 33 for the five elements most closely related to the Sigmodon L1s. Thus the elements that appeared to be most recently inserted of those characterized have accumulated ~6% divergence from their template sequence due to private mutations, suggesting that they inserted into the genome ~5.5 to 7.5 mya. In contrast, the most recently inserted Sigmodon and Peromyscus elements have accumulated fewer than 1% private mutations, suggesting that they inserted into the genome much more recently.
In the absence of transposition to create new elements, how long would it take to deplete the host genome of active templates? Hardies and Rikke (1989) have estimated the half-life of active L1s in rodents to be ~80,000 years. This estimate, 1/(neutral rate × length × fraction of strongly deleterious mutations), was based on a neutral mutation rate of 0.5% per million years, L1 sequence length of 5000 bp, and the assumption that 50% of mutations would be strongly deleterious. Correcting for the accepted neutral mutation rate in rodents of 1% per million years (She et al. 1990), this estimate would be 40,000 years (S. C. Hardies, personal communication), but a more conservative assumption that ~10% of mutations will be debilitating gives an estimated half-life of as much as 200,000 years. Thus the last detectable L1 activity in Oryzomys, on the basis of this data set, occurred >25 (given a half-life of 200,000 years) and perhaps >130 (given a half-life of 40,000 years) half-lives ago. Only about one in a million active elements would be surviving after 20 half-lives. If the number of potentially active elements in Oryzyomys before L1s became quiescent was ~3,000, similar to the number in Mus (DeBerardinis et al. 1998), it is unlikely that active templates are surviving from that last burst of L1 activity. It is more difficult to determine if L1s might be surviving by low rates of transposition. It may not be necessary that all individuals in a population contain active L1 masters in order for the element to survive, but it seems likely that they would at least contain signatures of recent transposition events.
L1s are the most persistent of mammalian transposable elements, dating back at least to the mammalian radiation. In contrast, SINE elements appear to have distinct origins in different groups of mammals—Alus are unique to primates and B1s unique to rodents, for example. High-copy number retrovirus-like elements such as IAP (Kuff et al. 1983), THE (McNaughton et al. 1995), and mys (Wichman et al. 1985) also appear to have a limited phylogenetic distribution in mammals. Why do L1s persist over such long periods? This apparent conservation has led to speculation that these elements provide an advantage to the genomes they inhabit. One of the more interesting suggestions is that they may serve to repair chromosomal breaks (Hutchison III et al. 1989), acting as a sort of “DNA band-aid.” This suggestion was made prior to the realization that L1 encodes its own endonuclease (Feng et al. 1996), but examination of insertion sites shows that not all L1s are inserted into the endonuclease target sequence. Thus, it is possible that some L1 insertions may be patching breaks in DNA.
The existence of retrotransposon-mediated repair has now been established experimentally (Moore and Haber 1996; Teng et al. 1996), but the significance of this pathway in natural systems remains an open question. If L1s mediate DNA break-repair, one might predict that if activity of L1 is low or absent, a species might be particularly susceptible to genetic or environmental agents that increase the incidence of chromosomal breaks. Interestingly, both Oryzomys and Holochilus have been found to have extremely unstable karyotypes in some natural populations. Koop et al. (1983) found that of 10 Oryzomys examined from a single isolated population, each animal had a different karyotype. Nachman and Myers (1989) have documented a similar phenomenon in Holochilus, where they found 26 distinct karyotypes among 42 animals trapped. Although these data are purely correlative, they are consistent with a possible role of L1 in DNA break-repair.
Even if L1s are not maintained in the host because of some positive function such as break-repair, their presence undoubtedly has shaped the mammalian genome. For example, it is thought that L1s provide both the reverse transcriptase and the endonuclease function for SINE retrotransposition and may be important for the formation of pseudogenes. The identification of a potential model mammalian system lacking L1 activity may provide an opportunity to examine the impact of L1 on mammalian genome organization in more detail.
Acknowledgement
We thank Nancy Moncrief, Priscilla Tucker, Michael Smith, and the Natural Science Research Lab of Texas Tech University for providing tissues for this study. We thank David Swofford for permission to use PAUP* prior to publication. This work was supported by a grant from the National Institutes of Health (R01-GM38737 to H.A.W.). L.E.W. was supported in part by a grant from the Howard Hughes Medical Institute through the Biological Sciences Undergraduate Education Program at Texas Tech University.
Footnotes
Communicating editor: W. F. Eanes
LITERATURE CITED
Author notes
Present address: Department of Agronomy, Iowa State University, Ames, IA 50011.
Present address: Medical Scientist Training Program, Baylor College of Medicine, 1 Baylor Plaza, Houston, TX 77030.