-
PDF
- Split View
-
Views
-
Cite
Cite
Carina Hethke, Agnes Bergerat, Winfried Hausner, Patrick Forterre, Michael Thomm, Cell-Free Transcription at 95°: Thermostability of Transcriptional Components and DNA Topology Requirements of Pyrococcus Transcription, Genetics, Volume 152, Issue 4, 1 August 1999, Pages 1325–1333, https://doi.org/10.1093/genetics/152.4.1325
Close - Share Icon Share
Abstract
Cell-free transcription of archaeal promoters is mediated by two archaeal transcription factors, aTBP and TFB, which are orthologues of the eukaryotic transcription factors TBP and TFIIB. Using the cell-free transcription system described for the hyperthermophilic Archaeon Pyrococcus furiosus by Hethke et al., the temperature limits and template topology requirements of archaeal transcription were investigated. aTBP activity was not affected after incubation for 1 hr at 100°. In contrast, the half-life of RNA polymerase activity was 23 min and that of TFB activity was 3 min. The half-life of a 328-nt RNA product was 10 min at 100°. Best stability of RNA was observed at pH 6, at 400 mm K-glutamate in the absence of Mg2+ ions. Physiological concentrations of K-glutamate were found to stabilize protein components in addition, indicating that salt is an important extrinsic factor contributing to thermostability. Both RNA and proteins were stabilized by the osmolyte betaine at a concentration of 1 m. The highest activity for RNA synthesis at 95° was obtained in the presence of 1 m betaine and 400 mm K-glutamate. Positively supercoiled DNA, which was found to exist in Pyrococcus cells, can be transcribed in vitro both at 70° and 90°. However, negatively supercoiled DNA was the preferred template at all temperatures tested. Analyses of transcripts from plasmid topoisomers harboring the glutamate dehydrogenase promoter and of transcription reactions conducted in the presence of reverse gyrase indicate that positive supercoiling of DNA inhibits transcription from this promoter.
PYROCOCCUS furiosus is an anaerobic marine Archaeon that grows optimally at 100° (Fiala and Stetter 1986). Because of their hyperthermophilic characteristics, members of the genus Pyrococcus are among the most-studied hyperthermophiles to date. Several Pyrococcus proteins have been shown to function at temperatures >90° in vitro (Leuschner and Antranikian 1995).
We recently described a cell-free transcription system for P. furiosus (Hethke et al. 1996). This system was used to identify the homologs of the eukaryotic transcription factors TFIIB and TBP that were detected in the genomes of Archaea (Ouzounis and Sander 1992; Marsh et al. 1994; Rowlands et al. 1994) as archaeal transcription factors (Hausner et al. 1996). Eukaryotic TBP is homologous to the archaeal transcription factor B (aTFB) and eukaryotic TFIIB is homologous to archaeal transcription factor A (aTFA), which have been purified from Methanococcus (Frey et al. 1990) and Pyrococcus cells (Hethke et al. 1996). To express their homology with eukaryotic transcriptional components, the factors aTFB and aTFA are now designated as archaeal TATA-binding protein (TBP) and TFB. aTBP and TFB interact similarly with promoter DNA as do their eukaryotic counterparts (Hausner and Thomm 1993; Nikolov et al. 1995). Although the basic pathway of promoter activation in Pyrococcus has been elucidated, the upper limit of cell-free transcription has been identified at a temperature 15° below the growth optimum of Pyrococcus (Hethke et al. 1996).
The topological state of plasmid DNA in hyperthermophilic Archaea varies with growth temperature from relaxed to positively supercoiled (Lopez-Garcia and Forterre 1997). Recently, linguistic analyses of complete genomes from hyperthermophilic Archaea also provided evidence for this unusual DNA topology (Herzel et al. 1998). Positive supercoiling of DNA might be caused by the activity of the enzyme reverse gyrase that exists only in hyperthermophiles (Forterre et al. 1995). Here, we report on an analysis of the limits of thermostability of all components involved in transcription and on the effects of DNA topology and reverse gyrase activity on archaeal cell-free transcription.
MATERIALS AND METHODS
Purification of transcriptional components: The superdex fractions of Pyrococcus RNA polymerase and recombinant Pyrococcus TFB and TBP were purified as described (Hausner et al. 1996). For the experiments shown in Figure 4, the superdex fraction of RNA polymerase was used for RNA synthesis. For all other experiments a more-purified RNA polymerase fraction was used, because the superdex fraction contained reverse gyrase activity (data not shown). A total of 1.7 ml of superdex fraction was concentrated by ultrafiltration (Centricon 100 Konzentrator; Amicon, Witten, Germany) to a volume of 40 μl diluted with 400 μl of a buffer containing 40 mm KPO4, pH 7.5, 1 mm EDTA, 1.5 m NaCl, 0.01% Triton X-100. This step was repeated once, and the enzyme in 440 μl was applied to a phenyl-Sepharose (CL-4B; Pharmacia, Uppsala, Sweden) column (0.6 × 7 cm) equilibrated with the same buffer. The column was washed with 10 ml of buffer and then with 10 ml of the same buffer containing only 0.5 m NaCl (flowrate 0.1 ml/hr). The enzyme was eluted from the column with the same buffer containing no salt but 10% (v/v) ethylene glycol in addition. In the phenyl-Sepharose fraction of RNA polymerase, reverse gyrase activity could not be detected.
Cell-free transcription reactions and preincubation experiments:
Assays for in vitro transcription from circular templates. These assays were conducted in a total volume of 50 μl in a buffer containing 40 mm HEPES, pH 6.5 (adjusted at 20°), 250 mm KCl, 2.5 mm MgCl2, 0.1 mm EDTA, 10 mm DTT, 1 μg template DNA (pLUW479), and 0.33 mm each of ATP, GTP, CTP, and UTP. Reaction mixtures contained 10 μl phenyl-Sepharose fraction of RNA polymerase and 0.2 μl each of aTBP and TFB. The molar ratio of template to RNA polymerase was ∼10:1 and the conditions allowed recycling of the RNA polymerase during the reactions. The assays were conducted in a PCR cycler (GeneAmp PCR system; Perkin-Elmer, Weiterstadt, Germany) using thin-walled tubes. To allow temperature adjustment, the reaction mixtures without ribonucleoside triphosphates (NTPs) were preheated for 1 min and the transcription started by addition of NTPs in a volume of 1.5 μl. After 10 min the reactions were stopped by shock-freezing in ethanol/dry ice. Transcripts were detected by primer extension in the presence of actinomycin D (50 μg/ml) as described previously (Hethke et al. 1996; Roovers et al. 1997).
Preincubation of proteins. All preincubation reactions (b-d) were conducted in the PCR cycler in thin-walled tubes. TBP, TFB, and RNA polymerase were preincubated in 25-μl transcription reactions [composition as described in (1)] not containing DNA and NTPs. Residual activity of preincubated proteins was determined after the addition of 25 μl buffer containing 1 μg template DNA (pLUW479 linearized with BamHI), 0.66 mm each of ATP, GTP, and CTP, 0.004 mm UTP, 2 μCi [α-32P]UTP (3000 Ci/mmol) and, if necessary, the protein components omitted from preincubation reactions. Transcription reactions were incubated for 30 min at 75°.
Preincubation of DNA. A total of 0.5 μg of circular DNA was incubated in a total volume of 25 μl in transcription reactions not containing TBP, TFB, RNA polymerase, and NTPs.
Preincubation of RNA. A 328-nt labeled run-off transcript from pLUW409 linearized with NdeI was produced and purified as described previously (Hethke et al. 1996). The RNA product was incubated in transcription buffer not containing DNA, NTPs, or proteins in a total volume of 20 μl. The pH and the salt and Mg2+ concentrations were varied as indicated in the figure legends.
Preincubation of NTPs. A total of 0.66 mm each of ATP, GTP, CTP, 0.004 mm UTP, and 2 μCi [α-32P]UTP were incubated in 25-μl transcription reactions not containing protein components and DNA.
Preparation of topoisomers: Negatively supercoiled (--) plasmid DNA was prepared from Escherichia coli cells by CsCl centrifugation. Slightly negatively supercoiled DNA (-) was prepared from (--) DNA by treatment with eukaryotic DNA topoisomerase I in the presence of ethidium bromide. A total of 30 μg pLUW479-DNA was incubated in a total volume of 300 μl in a buffer containing 50 mm Tris-HCl (pH 8.0), 100 mm KCl, 5 mm MgCl2, 0.5 mm DTT, 0.1 mm EDTA, 100 μg/ml BSA, and 2 μg/ml ethidium bromide. This mixture was preincubated for 10 min at 37°, and then 50 units of topoisomerase I were added. After 90 min at 37°, the reaction was stopped by the addition of 30 μl 10% (w/v) SDS. Ethidium bromide was removed by repeated extraction with n-butanol.
Relaxed plasmid DNA (o) was obtained by incubation of 20 μg pLUW479-DNA with 500 units of topoisomerase I of E. coli. The reaction mixture contained, in a final volume of 200 μl, 35 mm Tris-HCl, pH 8.0, 20 mm KCl, and 2 mm MgCl2. The reaction was incubated for 90 min at 37°. The reaction was stopped by the addition of 20 μl of a 10% (w/v) SDS solution.
Positively supercoiled DNA was prepared by incubating 9 μg DNA (--) with 50 units of reverse gyrase from P. furiosus (Borges et al. 1997) in a total volume of 200 μl. The buffer contained 50 mm Tris-HCl, pH 8.8, 150 mm NaCl, 10 mm MgCl2, 1 mm ATP, 1 mm EDTA, 2 mm DTT, and 250 μg/ml BSA. To obtain slightly positively supercoiled DNA (+), the reaction was incubated for 2 min and 15 sec at 90°, and for the preparation of positively supercoiled (++) DNA at 100°. All topoisomers were purified by the use of QIAGEN (Hilden, Germany) Tip 20 columns, and the DNA eluted from the columns was precipitated with 2-propanol.
Determination of the superhelical densities of the different topoisomers was performed as described (Marguet and Forterre 1994) by analysis on 2D-gels containing various concentrations of chloroquine or netropsin in the first and second dimension.
RESULTS
Stability of components of the cell-free system: To address the question of whether the organic buffer substance and nucleotides are stable at high temperatures, the HEPES buffer and NTPs were preincubated in cell-free transcription reactions, the protein components and template DNA were added later, and RNA synthesis was measured. A preincubation of HEPES and NTPs for 30 min at 90° or 100° did not affect a subsequent transcription reaction (data not shown). This finding indicates that degradation of NTPs and of the HEPES buffer is not a factor limiting cell-free RNA synthesis at 100°.
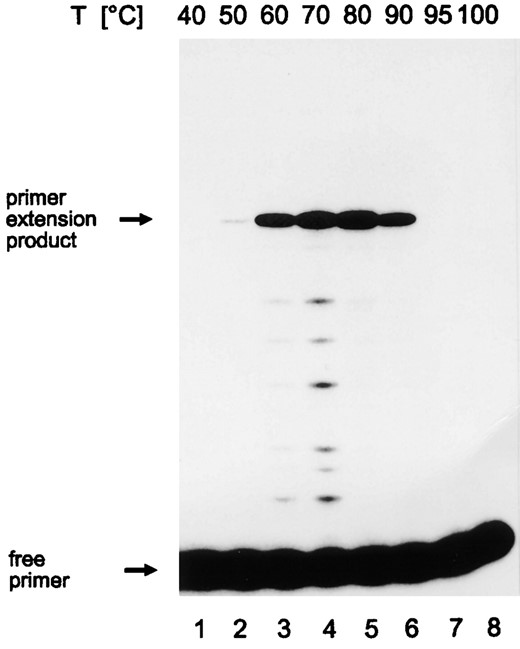
Analysis of run-off transcripts from linearized DNA templates had shown that a temperature of 85° represented the upper limit of cell-free transcription by the Pyrococcus RNA polymerase (Hethke et al. 1996), most likely due to denaturation of the linear DNA at temperatures >85°. As supercoiled DNA was reported to show considerably higher thermostability (Marguet and Forterre 1994), a primer extension assay allowing analysis of transcripts from circular templates was established. Analysis of RNA products transcribed from a circular negatively supercoiled plasmid encoding the Pyrococcus gdh gene revealed that effective RNA synthesis occurred in a temperature range between 60° and 90° (Figure 1, lanes 3-6). No transcripts were observed when the transcription reaction was incubated at 95° or 100° (Figure 1, lanes 7 and 8).
—Temperature optimum of Pyrococcus in vitro transcription from a circular template containing the gdh promoter. Cell-free transcription reactions were conducted at the temperatures indicated at the top. RNA synthesis was analyzed by primer extension. The amount of extension product was directly proportional to the concentration of template RNA when 1/16, 1/8, 1/4, and 1/2 of the RNA were synthesized in one standard cell-free transcription reaction and when RNA of one complete reaction were used as template. The positions of the 94-nt primer extension product and of the free primer are indicated. Primer extension products were separated on an 8% denaturing polyacrylamide (PA) gel.
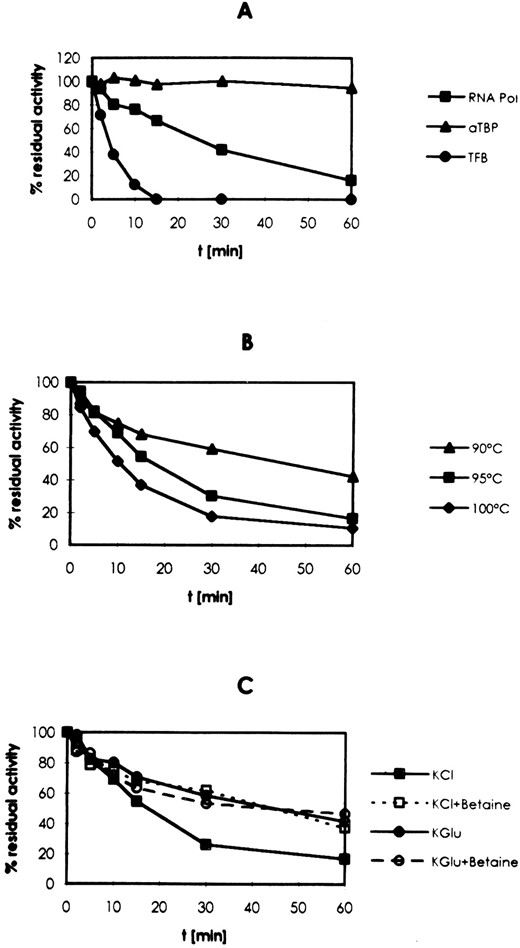
To identify the components of the transcriptional machinery limiting in vitro transcription at the optimal growth temperature of Pyrococcus, the RNA polymerase and the factors TFB and aTBP were preincubated at 100°, and their remaining activity was analyzed in cell-free transcription reactions. The activity of aTBP was not altered after 1 hr of incubation, whereas the activity of TFB decreased rapidly. The half-life time for TFB activity was 3 min. After 15 min at 100°, the activity of TFB was completely destroyed (Figure 2A). The half-life time of RNA polymerase activity was 23 min at this temperature. When aTBP, TFB, and RNA polymerase were preincubated in combination, the half-life time of the activity of the preinitiation complex was 44 min at 90° and 12 min at 100°. Even after 1 hr at 100°, ∼10% residual activity was observed (Figure 2B). This finding indicates that TFB is stabilized in the presence of aTBP and RNA polymerase.
The proteins can also be stabilized by an increase in the ionic strength or addition of betaine. When 1 m betaine was added to preincubation reactions or when 250 mm KCl was replaced by 400 mm K-glutamate, the thermostablity of the preinitiation complex was increased significantly (Figure 2C).
—Activity of aTBP, TFB, and RNA polymerase after preincubation at extreme temperatures. Proteins were incubated individually (A; 100°) or in combination (B and C) in 25-μl transcription reactions (A-C; materials and methods) not containing DNA and NTPs. After preincubation at the temperatures indicated, the activity of the components was determined in transcription reactions. (C) Preincubation was performed at 95°. The concentrations of salts and betaine used for preincubation reactions were as follows: 250 mm KCl (KCl), 1 m betaine (betaine), 400 mm K-glutamate (KGlu). RNA products separated as indicated in Figure 1 were quantitated by densitometry.
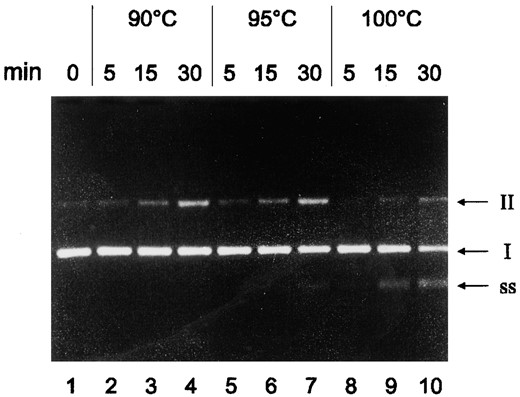
To investigate the thermostabilty of DNA under the conditions of cell-free transcription reactions, a negatively supercoiled plasmid harboring the Pyrococcus gdh-promoter was incubated at different temperatures and the DNA was analyzed after electrophoresis in agarose gels. When DNA was incubated for 15 and 30 min at 90° and 95°, the number of molecules containing nicks in the DNA was increased (Figure 3, lanes 2-4 and lanes 5-7). After 30 min at 95° or 15 and 30 min at 100°, an additional DNA band was observed that migrated faster than cccDNA (Figure 3, lanes 7 and 9). This DNA band most likely represents ssDNA as it could be degraded by nuclease S1 and could renature to open circular DNA (data not shown). In contrast to proteins of the transcriptional machinery, the DNA could not be stabilized by addition of betaine or by replacement of KCl with K-glutamate (data not shown).
—Thermostability of template DNA. The plasmid pLUW479, harboring the Pyrococcus gdh promoter, was incubated at different temperatures in transcription reactions not containing NTPs as indicated at the top. DNA was subsequently analyzed by electrophoresis on 10% agarose gels and stained with ethidium bromide. I, cccDNA; II, ocDNA; SS, single-stranded DNA.
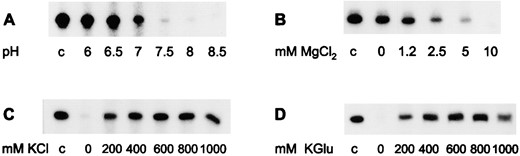
Stability of RNA: To investigate the conditions preventing hydrolysis of RNA synthesized in cell-free transcription reactions, the effect of pH and various salt conditions on the stability of labeled in vitro RNA was studied. RNA was found to be most stable at lower pH values. When the RNA was incubated in standard transcription reaction buffer (2.5 mm MgCl2, 250 mm KCl) at pH 7.5, the 328-nt run-off transcript was almost completely degraded after incubation at 90° for 30 min. At pH 6 and 6.5 >50% of RNA was still intact under the same conditions (Figure 4A).
Bivalent metal ions are known to catalyze hydrolysis of RNA and NTPs (Tetas and Lowenstein 1963; Lindahl 1967), but are required for the activity of most enzymes involved in synthesis and modification of DNA and RNA. To analyze the stability of RNA in the range of Mg2+ concentration required for cell-free transcription, labeled in vitro RNA products were incubated at pH 7 and 250 mm KCl in the presence of differing MgCl2 concentrations. Mg2+ cations were found to destabilize RNA dramatically. At 10 mm MgCl2, the RNA was completely degraded after 30 min at 90°, whereas >50% of the RNA was still detected in the absence of MgCl2 (Figure 4B). At 2.5 mm MgCl2 and pH 7, the percentage RNA remaining was ∼25 (Figure 4B) at the same MgCl2 concentration and at pH 6.5, ∼50 (data not shown). We found that a MgCl2 concentration of 2.5 mm at pH 6.5 was sufficient for RNA polymerase activity (data not shown) and RNA stability (Figure 4B).
—Effect of pH, Mg2+ ions, and salt on RNA stability at extreme temperatures. A total of 8 fmol of labeled in vitro RNA was incubated for 30 min at 90° in transcription reaction buffer. The pH of the buffer (A), the MgCl2 concentration (B), and the KCl concentration (C) were modified as indicated below the lanes. (D) KCl was replaced by K-glutamate; the pH was adjusted at 20°. After preincubation, the decrease of the 328-nt RNA product was analyzed on 6% denaturing PA gels.
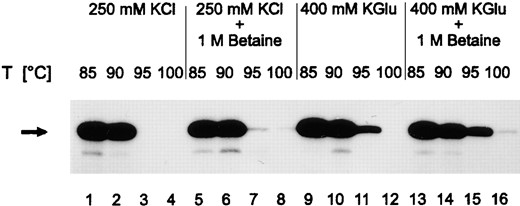
—Cell-free transcription at temperatures close to the boiling point of water. Transcription reactions were conducted for 10 min at different temperatures and contained salts and betaine as indicated on top of the lanes. Transcripts from circular templates were identified by primer extension. cDNA was analyzed on 8% denaturing PA gels. The arrow indicates the major primer extension product. The shorter products originate from premature stop of primer extension reactions.
To identify the ionic conditions favoring RNA stability at extreme temperatures, labeled RNA was incubated in standard transcription reaction buffer (pH 6.5; 2.5 mm MgCl2) at different concentrations of KCl and K-glutamate. In the absence of KCl, RNA was almost completely degraded after 30 min at 90°. At a salt concentration of 200 mm KCl or K-glutamate, the stability of RNA was increased, and optimal stability was observed in a range between 400 and 800 mm (Figure 4, C and D). The KCl optimum of cell-free transcription was found at 250 mm. At 400 mm KCl, no RNA synthesis was observed (data not shown). In contrast, effective RNA synthesis was still possible at a K-glutamate concentration of 400 mm (data not shown and Figure 5). These findings suggest that transcription is not inhibited by K+ cations up to 400 mm but is inhibited by Cl- anions at the same concentration. The data shown in Figure 4, C and D, indicate that potassium ions stabilize RNA at concentrations between 200 and 800 mm. Kinetic analyses confirmed this conclusion. At 90° the half-life of the 328-nt RNA products was 20 min in the presence of 250 mm KCl and 25 min in the presence of 400 mm K-glutamate. The addition of betaine resulted in a further stabilization of RNA. The half-life of RNA at 90° was 33 min in the presence of 400 mm K-glutamate and 1 m betaine (data not shown).
Optimization of transcription at 95°: As the inactivation of protein components of the transcriptional machinery is lower and the stability of RNA is higher in the presence of K-glutamate and of betaine, transcription reactions were conducted in the presence of these compounds. The 94-nt RNA product transcribed in 10 min from negatively supercoiled DNA was analyzed by a primer extension reaction (materials and methods). The products analyzed in Figure 5 are the result of RNA synthesis and of product decay at a given temperature. The latter is increased at higher temperatures (see below). When 1 m betaine was added to cell-free transcription reactions containing 250 mm KCl, a weak RNA product could still be detected after incubation at 95° (compare Figure 5, lanes 3 and 7). When 400 mm K-glutamate was added to transcription reactions carried out at 95°, the amount of RNA detected by primer extension was clearly increased (Figure 5, lane 11). In the presence of both 400 mm K-glutamate and 1 m betaine, much more RNA was found at 95° (Figure 5, lane 15), and even in a transcription reaction conducted at 100°, a weak signal was observed (Figure 5, lane 16). Higher concentrations of betaine, known to stabilize proteins to higher extent (Tetas and Lowenstein 1963), were found to inhibit cell-free transcription (data not shown).
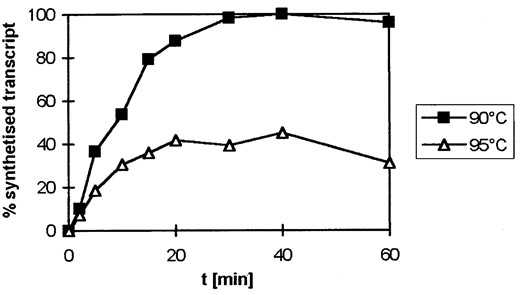
To extend these analyses, the kinetics of cell-free transcription were analyzed at 100°, 95°, and 90° in the presence of K-glutamate and 1 m betaine. At 100°, it was not possible to demonstrate an increase of specific transcripts depending on the incubation time. But at 90° and 95°, an increase of RNA product with incubation time was observed (Figure 6). The increase of RNA product detected was quasilinear for 16 min at 90° and 10 min at 95°. These findings demonstrate that cell-free RNA synthesis by the Pyrococcus RNA polymerase is possible up to a temperature of 95°. Although 60% of transcriptional activity at 95° is retained after 30 min preincubation of the protein components of the transcriptional machinery in the presence of K-glutamate and betaine (Figure 2C), there is no further accumulation of product after 30 min (Figure 6). At least two processes lead to the plateau shown in Figure 6: enzyme inactivation, which seems to be minor, and product decay, which seems to be major. To analyze the extent of product decay directly, radiolabeled RNA (328 nt) was incubated in the buffer conditions used in Figure 6 and the half-life of RNA at 90°, 95°, and 100° was analyzed. Under these conditions, the half-life of the 328-nt RNA product was 33 min at 90°, 20 min at 95°, and 10 min at 100° (data not shown). If decay is a random fragmentation process, the half-life of the 94-nt RNA product analyzed in Figure 6 would be longer because of the smaller target size of this molecule. Considering its smaller size, the half-life of this 94-nt RNA is 115 min at 90°, 69 min at 95°, and 35 min at 100°. These data show clearly that RNA decay affects product accumulation analyzed in Figure 6 more at higher temperatures, but RNA decay alone is not responsible for the difference between the two curves in Figure 6. The initial slopes of the hyperbolic curves in Figure 6 are similar, suggesting that initial enzyme activity at 90° and 95° is also similar, but besides RNA decay, other processes like inactivation of the DNA template seem to be responsible for the lower amount of product accumulation found at 95°. As the RNA half-life of the 94-nt RNA product at 100° is still 35 min, the failure to demonstrate accumulation of an RNA product at 100° after a 10-min transcription reaction (Figure 5) is most likely caused by inactivation of transcription factors and template DNA at this temperature.
—Kinetics of in vitro transcription at 90° and 95°. Transcription reactions were conducted in the presence of 400 mm K-glutamate and 1 m betaine. RNA products were identified by primer extension. Labeled cDNA was separated on 8% denaturing PA gels, and autoradiograms were quantified by densitometry.
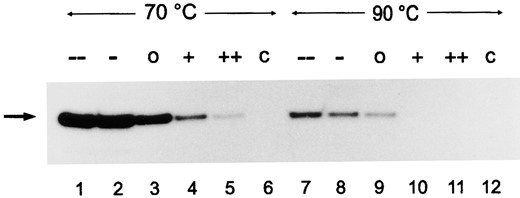
Effect of DNA topology on cell-free transcription: To analyze the requirements for template topology in the Pyrococcus cell-free system, a range of plasmid topoisomers containing the Pyrococcus gdh promoter were prepared. The superhelical density of these plasmids ranged at 25° from negatively supercoiled (σ = -0.052) to positively supercoiled (σ = +0.029; Table 1). A set of topoisomers representing negatively supercoiled DNA (--), slightly negatively supercoiled DNA (-), relaxed DNA (o), slightly positively supercoiled DNA (+), and positively supercoiled DNA (++) were used as templates for cell-free transcription reactions at 70° and 90°. At both temperatures, negatively supercoiled DNA was clearly the best template. At 70°, relaxed and slightly positively supercoiled DNA were transcribed efficiently and even positively supercoiled DNA was transcribed weakly. At 90°, the transcriptional activity in general was lower. Positively supercoiled DNA was not transcribed, and slightly positively supercoiled DNA was transcribed weakly (Figure 7; longer exposure of the autoradiogram not shown). Negative supercoiling of DNA decreases with increasing temperature and positive supercoiling increases (Charbonnier et al. 1992; Table 1). Considering this temperature dependence of DNA supercoiling, cell-free transcription was possible from DNA topoisomers in a range of σ = -0.039 to +0.043 at 70° and in a range from σ = -0.034 to +0.032 at 90°. The topology of the sets of plasmids used as templates for transcription reactions was not altered after transcription of the templates. Analysis of DNA from transcription reactions in agarose gels revealed only a weak increase of nicked DNA molecules (data not shown).
Superhelical densities of topoisomers of pLUW479 at different temperatures
| Plasmid DNA . | σ at 25° . | σ at 70° . | σ at 90° . |
|---|---|---|---|
| (--) | -0.052 | -0.039 | -0.034 |
| (-) | -0.027 | -0.015 | -0.009 |
| (o) | -0.005 | +0.018 | +0.024 |
| (+) | +0.013 | +0.026 | +0.032 |
| (++) | +0.029 | +0.043 | +0.049 |
| Plasmid DNA . | σ at 25° . | σ at 70° . | σ at 90° . |
|---|---|---|---|
| (--) | -0.052 | -0.039 | -0.034 |
| (-) | -0.027 | -0.015 | -0.009 |
| (o) | -0.005 | +0.018 | +0.024 |
| (+) | +0.013 | +0.026 | +0.032 |
| (++) | +0.029 | +0.043 | +0.049 |
The linking number of the major topoisomers was used for calculation of σ values of (--), (o), and (++) pLUW479 DNA. For the calculation of the σ values of (-) and (+) pLUW479 DNA, the average linking numbers of the most positive and most negative topoisomer of each preparation were used. The superhelical densities are given with an error margin of ±0.003.
Superhelical densities of topoisomers of pLUW479 at different temperatures
| Plasmid DNA . | σ at 25° . | σ at 70° . | σ at 90° . |
|---|---|---|---|
| (--) | -0.052 | -0.039 | -0.034 |
| (-) | -0.027 | -0.015 | -0.009 |
| (o) | -0.005 | +0.018 | +0.024 |
| (+) | +0.013 | +0.026 | +0.032 |
| (++) | +0.029 | +0.043 | +0.049 |
| Plasmid DNA . | σ at 25° . | σ at 70° . | σ at 90° . |
|---|---|---|---|
| (--) | -0.052 | -0.039 | -0.034 |
| (-) | -0.027 | -0.015 | -0.009 |
| (o) | -0.005 | +0.018 | +0.024 |
| (+) | +0.013 | +0.026 | +0.032 |
| (++) | +0.029 | +0.043 | +0.049 |
The linking number of the major topoisomers was used for calculation of σ values of (--), (o), and (++) pLUW479 DNA. For the calculation of the σ values of (-) and (+) pLUW479 DNA, the average linking numbers of the most positive and most negative topoisomer of each preparation were used. The superhelical densities are given with an error margin of ±0.003.
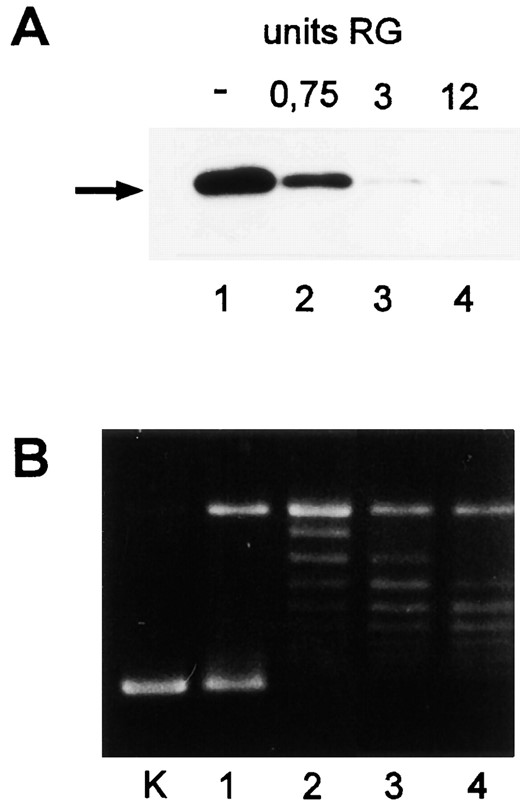
To study the DNA topology dependence of cell-free transcription in more detail, reverse gyrase was added to transcription reactions containing negatively supercoiled plasmid DNA (--). Reverse gyrase activity inhibited transcription strongly. A total of 0.75 units of this enzyme reduced transcription significantly; in the presence of 3 and 12 units, only residual RNA synthesis was observed (Figure 8A, lanes 2-4). Reverse gyrase was active in standard cell-free transcription reactions. It increased the amount of relaxed DNA when 0.75 units were added (Figure 8B, lanes 1 and 2). Positively supercoiled topoisomers were formed at higher concentrations (Figure 8B, lanes 3 and 4 and data not shown). These findings indicate that the activity of reverse gyrase and positive supercoiling of DNA inhibit transcription from the gdh promoter by the Pyrococcus RNA polymerase.
—Effect of DNA topology on cell-free transcription. Plasmid pLUW479 in different conformations, negatively supercoiled (--), slightly negatively supercoiled (-), relaxed (o), slightly positively supercoiled (+), and positively supercoiled (++) was used as template in cell-free transcription reactions, and the RNA was analyzed by primer extension. C is a control reaction not containing NTPs in transcription reactions.
—Reverse gyrase activity inhibits transcription. The units of reverse gyrase added to transcription reactions are indicated on top of the lanes. The initial template was negatively supercoiled DNA (-). (A) Detection of transcripts by primer extension and analysis of cDNA on a denaturing 8% PA gel. Primer extension products are labeled by an arrow. (B) Analysis of reverse gyrase activity. Aliquots of the transcription reactions containing 0.25 μg DNA were analyzed on 1% agarose gels, and the topoisomers were stained with ethidium bromide after electrophoresis. K, negatively supercoiled pLUW 479 DNA.
DISCUSSION
P. furiosus grows in a temperature range between 70° and 103° (Fiala and Stetter 1986). Cell-free transcription occurred on circular DNA as a template in transcription assays between 50° and 90° (Figure 1). As the upper limit of cell-free transcription was below the optimal growth temperature of the organism, it is obvious that the transcriptional components showed lower thermostability in vitro than in Pyrococcus cells. To define the upper temperature limit of cell-free transcription is important both for understanding the biology of transcription of a hyperthermophile and for biotechnological applications in general, as in vitro synthesis of nucleic acids at high temperatures becomes increasingly important in molecular biology techniques.
Surprisingly, the stability of NTPs was not altered significantly even after incubation at 100° for 30 min. This is in contrast with the low half-life time in the range of minutes reported for ATP (Stetter et al. 1986; Phipps et al. 1991). Phipps et al. (1991) used a buffer containing 9 mm MgCl2 as a single salt for preincubation experiments. As high concentrations of Mg2+ ions accelerate degradation of RNA and high concentrations of K+ ions stabilize RNA (Figure 4), this difference in thermostability is most likely caused by the presence of 250 mm KCl and the lower Mg2+ concentration (2.5 mm) in the cell-free transcription reactions.
The residual activity of protein components after preincubation at extreme temperatures was different. aTBP, like many proteins from hyperthermophiles, which show half-life times of several hours at 100° (Adams 1993; Kengen et al. 1993), did not lose any activity after 1 hr at 100° (Figure 2A). The finding that RNA polymerase lost only 50% of its activity after 23 min at 100° (Figure 2A) and was able to synthesize RNA for a period of 20 min at 95° (Figure 6) indicates that even this enzyme, with a molecular mass of ∼500,000, consisting of 11 different subunits, shows a high intrinsic thermostability. In contrast, TFB was inactivated almost completely after 10 min at 100° (Figure 2A). As the residual activity of the transcription system was still 20% after 30 min at 100°, when TFB was preincubated in combination with TBP and RNA polymerase (Figure 2B), TFB seemed to be stabilized by protein-protein contacts. Additional extrinsic factors may confer thermal stability to TFB in Pyrococcus cells.
The intracellular K+ concentration in Pyrococcus cells is 500-600 mm (Scholz et al. 1992). We demonstrate here that physiological concentrations of K+ ions stabilize RNA (Figure 4) and increase the temperature maximum of transcription from 90° to 95° (Figure 5). Our results show that mRNA is most stable in vitro between pH 6.0 and 6.5 in the absence of Mg2+ ions and identify K+ ions clearly as an extrinsic factor contributing to stability and enzymatic activity of Pyrococcus transcriptional components. The organic solute di-myo-inositol-phosphate is accumulated in cells of Pyrococcus in response to temperature (Martins and Santos 1995) and was shown to stabilize proteins against thermal denaturation (Scholz et al. 1992). This compound, however, was found here to inhibit cell-free transcription (B. Goede and M. Thomm, unpublished data), most likely due to binding to DNA in vitro (E. Marguet and P. Forterre, unpublished data). Therefore, this intracellular solute is not suitable to improve the efficiency of cell-free transcription reactions at extreme temperatures. However, the compatible solute betaine, which does not exist in Pyrococcus cells, increased the activity of the cell-free system at 95° significantly (Figure 5). Betaine is known to shift the thermal unfolding transition temperature (Tm) of proteins to higher temperatures (Santoro et al. 1992) and did not inhibit transcription up to a concentration of 1 m. Therefore, this compound seems to be an excellent additive for cell-free polymerization reactions carried out at extreme temperatures.
The system described here allows cell-free RNA synthesis from supercoiled templates at temperatures very close to the boiling point of water. Cell-free transcription reactions can be carried out with high activity at 90° and with still significant activity at 95°. The upper limit of transcription of a cell-free system established for the hyperthermophilic Bacterium Thermotoga and for the Crenarchaeote Sulfolobus was at 75° (Hüdepohl et al. 1990; Meier et al. 1995). The Pyrococcus transcriptional components are the most thermostable ones reported, and Pyrococcus cell-free systems may therefore prove to be ideal model systems to study macromolecule synthesis at extreme temperatures.
In contrast to the DNA of mesophilic Archaea and of Bacteria that is negatively supercoiled (σ = -0.05 to -0.07), the superhelicity of plasmids from hyperthermophiles ranges from relaxed to positively supercoiled (σ = -0.013 to +0.035; Forterre et al. 1995). In this study, we established a reconstituted Pyrococcus cell-free transcription system from bacterially produced TFB and aTBP and a highly purified phenyl-Sepharose fraction of RNA polymerase that, in contrast to all hitherto used preparations of this enzyme, was free of reverse gyrase activity and therefore suitable to investigate the DNA topology requirements of transcription. The DNA topoisomers purified as templates for cell-free transcription experiments range in their superhelicity from -0.04 to +0.05 (Table 1). Our findings presented here show that positively supercoiled DNA can be transcribed in vitro both at 70° (σ = +0.018 to +0.043) and at 90° (σ = +0.024 to +0.032). These results therefore provide evidence that naturally occurring positively supercoiled DNA can be transcribed by the Pyrococcus transcriptional machinery. As the experiments were conducted with highly purified components, our data also provide evidence that additional factors like helicases are not required for separation of DNA strands at the promoter even when the DNA is in the energetically less favorable positive superhelical conformation. In addition, hydrolysis of the β-γ-bond of ATP was not required for initiation of transcription from linear archaeal templates (W. Hausner and M. Thomm, unpublished data). These findings are of particular interest as the archaeal transcription system is closely related to the eukaryotic RNA polymerase II system, which requires the hydrolysis of the β-γ bond of ATP (Dvir et al. 1996) and the helicase activity of TFIIH for open complex formation (Kugel and Goodrich 1998). However, analyses of transcripts from topoisomers (Figure 7) and of RNA synthesis in the presence of reverse gyrase (Figure 8) show clearly that negatively supercoiled DNA is the preferred template compared with relaxed DNA and that positive supercoiling deteriorates the template activity of DNA (Figures 7 and 8). Analyses of eukaryotic and bacterial transcription systems provided similar results. In general, the efficiency of transcription from positively supercoiled DNA was found to be extremely low, from relaxed DNA to be low (Bird et al. 1992; Gartenberg and Wang 1992), and negative supercoiling of the template was found to stimulate the efficiency of transcription. In contrast, template topology of an archaeal rRNA promoter has negligible effect on RNA synthesis at physiological temperatures in a cell-free Sulfolobus transcription system (Bell et al. 1998). Although it is unclear whether this difference depends on the type of promoter or on a particular property of the Sulfolobus system, the data reported here provide the first evidence that negatively supercoiled DNA is also the preferred template in archaeal transcription. Future analyses of different Pyrococcus promoters, e.g., those for genes encoding DNA topoisomerases, which are known to be regulated in response to DNA topology in Bacteria (Menzel and Gellert 1983; Tse-Dinh 1985), will show to what extent DNA topology is involved in regulation of gene transcription in Archaea.
Acknowledgement
We thank Erwin Galinski, Universität Münster, for valuable discussions and providing di-myo-inositol-phosphate and Bernd Goede, Universität Kiel, for investigating the effect of this compound on cell-free transcription. We appreciate the support of this work provided by Evelyne Marguet. This work was supported by grants of the Biotechnology program of the European Community to M. Thomm and P. Forterre. The work of M. Thomm was funded by the “Fonds der Chemischen Industrie.”
Footnotes
Communicating editor: C. J. Daniels
LITERATURE CITED