-
PDF
- Split View
-
Views
-
Cite
Cite
Kevin F O'Connell, Charles M Leys, John G White, A Genetic Screen for Temperature-Sensitive Cell-Division Mutants of Caenorhabditis elegans, Genetics, Volume 149, Issue 3, 1 July 1998, Pages 1303–1321, https://doi.org/10.1093/genetics/149.3.1303
Close - Share Icon Share
Abstract
A novel screen to isolate conditional cell-division mutants in Caenorhabditis elegans has been developed. The screen is based on the phenotypes associated with existing cell-division mutations: some disrupt postembryonic divisions and affect formation of the gonad and ventral nerve cord—resulting in sterile, uncoordinated animals—while others affect embryonic divisions and result in lethality. We obtained 19 conditional mutants that displayed these phenotypes when shifted to the restrictive temperature at the appropriate developmental stage. Eighteen of these mutations have been mapped; 17 proved to be single alleles of newly identified genes, while 1 proved to be an allele of a previously identified gene. Genetic tests on the embryonic lethal phenotypes indicated that for 13 genes, embryogenesis required maternal expression, while for 6, zygotic expression could suffice. In all cases, maternal expression of wild-type activity was found to be largely sufficient for embryogenesis. Cytological analysis revealed that 10 mutants possessed embryonic cell-division defects, including failure to properly segregate DNA, failure to assemble a mitotic spindle, late cytokinesis defects, prolonged cell cycles, and improperly oriented mitotic spindles. We conclude that this approach can be used to identify mutations that affect various aspects of the cell-division cycle.
TO divide, a cell must be able to replicate its DNA, assemble and position a mitotic spindle, and initiate and complete a cytokinetic furrow at the appropriate time and place. Each of these events and all the associated intermediate steps must be coordinated with one another and executed with a high degree of precision to allow the faithful segregation of genetic material and cytoplasmic constituents. Despite a long-standing effort to elucidate the mechanisms involved, a complete understanding of many aspects of the process of cell division is lacking.
Using mutation to identify genes required for cell division is one particularly fruitful approach to understanding these mechanisms. Mutant hunts in budding yeast (Hartwell et al. 1970), fission yeast (Nurse et al. 1976; Nasmyth and Nurse 1981; Samejima et al. 1993), flies (Gatti and Baker 1989), and other species have led to the identification of many components of the cell-division machinery. But understanding in molecular terms the function of an individual component and how it interacts with other essential proteins requires that genetics be combined with biochemistry and cell biology. Thus, the ideal biological system should be amenable not only to genetic studies but also to a broad range of biochemical and cell biological techniques.
The nematode Caenorhabditis elegans seems well suited to exploring the mechanisms of cell division. In addition to its availability for strong genetic techniques, C. elegans has proven useful for both biochemical (Aroian et al. 1997) and cell biological (Hyman 1989; Hird and White 1993) approaches. The early embryo, in particular, possesses several attractive features for such studies: it is immotile and contains fairly large cells (10 to 50 μm in diameter), facilitating cytological analysis. The centrosomes and spindles are visible by light microscopy and can thus be viewed in live specimens without the need for complex imaging techniques. Equally important, cells in the C. elegans embryo undergo cytokinesis—unlike yeast cells, which divide by medial fission or budding, or early Drosophila embryos, which undergo nuclear division without cytokinesis. Thus, mutations that affect this aspect of cell division can be studied during the earliest stages of embryogenesis. In addition, the C. elegans embryo possesses both cells that divide proliferatively—to produce identical daughters—and cells that divide determinatively—to produce daughters with distinct developmental potentials. Thus, mutations that affect one or both types of division can be identified.
Mutations that affect cell division have been previously identified in C. elegans. However, the effects of many of these are limited to a subset of lineages. Several large-scale screens for embryonic lethal (emb) mutants have identified mutations that affect various aspects of cell division in the early embryo (Hirsh and Vanderslice 1976; Miwa et al. 1980; Cassada et al. 1981; Kemphues et al. 1988a,b), while another screen has identified mutations that specifically affect the postembryonic lineages (Horvitz and Sulston 1980). Both sets include mutations that disrupt the temporal and spatial patterns of cell division as well as mutations that block division altogether. As not all lineages are affected by these mutations, it is not known whether these genes encode key players in the mechanisms of division that are common to all cells.
Among the mutations that affect postembryonic lineages, lin-5(e1348) and lin-6(e1466) are unusual in that they affect nearly all divisions that occur during this period (Albertson et al. 1978; Horvitz and Sulston 1980; Sulston and Horvitz 1981). These two mutations affect very different aspects of the cell-division process; lin-5 mutants exhibit defective karyokinesis and cytokinesis (Albertson et al. 1978), while lin-6 mutants are defective in DNA synthesis (Sulston and Horvitz 1981). Despite the severity of the defects, both lin-5 and lin-6 homozygotes are viable; none of the cell divisions that occur postembryonically are essential for growth and development. However, both mutants exhibit a similar sterile, uncoordinated (Stu) phenotype because the cell divisions required for formation of the gonad and certain ventral-cord motor neurons fail.
It is possible that the lin-5 and lin-6 genes encode proteins that play fundamental roles in cell division. One would therefore expect that these genes would be required for all divisions. However, neither lin-5(e1348) nor lin-6(e1466) affects the embryonic divisions; homozygotes complete embryogenesis normally. A likely explanation for the lack of early defects could be that these gene products are maternally provided. Homozygotes would exhibit cell-division failure only when the maternal gene product becomes limiting—in these cases, during the postembryonic divisions. As the lin-5 and lin-6 mutants are nonconditionally sterile, they provide no insight into embryonic functions. One could, however, identify conditional alleles. This would allow one to block maternal gene expression and study the cytological phenotype during the embryonic divisions.
We have devised a screen to identify temperature-sensitive (ts), nonlineage-specific cell-division mutations in C. elegans. Our approach was designed with two goals in mind: to identify genes that encode key components of the cell-division machinery, and that are thus required for most cell divisions, and to be able to study the effects of these mutations during the early embryonic divisions. To identify the desired genes, we screened for mutants with ts Emb and Stu phenotypes, indicative of embryonic and postembryonic cell-division failures. Here we describe the results of this new approach, including genetic and cytological analysis for the 19 mutants isolated.
MATERIALS AND METHODS
Culture conditions and strains: All strains were cultured using standard techniques on nematode growth medium (NGM) seeded with Escherichia coli strain OP50 (Brenner 1974; Lewis and Fleming 1995). Temperature-sensitive mutants were grown at either 16° (permissive temperature) or 25° (restrictive temperature) and most other strains at 20°.
All strains were derived from the wild-type Bristol strain N2. The following mutations, descriptions of which can be found in Hodgkin et al. (1988), Hodgkin (1997), or in cited references, were used: LGI:dpy-5(e61), fer-1(hc13) (Ward and Miwa 1978), sem-2(n1343), mex-3(zu155), let-541(h886) (McKim et al. 1992), let-536(h882) (McKim et al. 1992); LGII:mel-11(it26), mel-9(b293), let-240(mn209), let-31(mn31), zyg-1(b1), dpy-10(e128), let-251(mn95), mel-15(it38), mel-22(it30), rol-1(e91), lin-5(e1348), let-266(mn194), spe-8(hc40), let-239(mn93), let-23(sy10), let-252 (mn100), let-19(mn19), let-238 (mn229), evl-20(ar103); LGIII: unc-32(e189), plg-1(e2001), ooc-4(e2078), cul-1(e1756) (Kipreos et al. 1996); LGIV: emb-3(hc59), let-54(s44), mes-6(bn66), gon-1(q518), gon-4(e2575), mel-24(ct59), emb-26(g47), emb-31(g55), unc-5(e53), let-296(s1250), let-292(s1146), let-297(s1989), let-71(s692), let-64(s216), let-73(s685), let-655(s1748), evl-7(ar108), fem-1(e1991), spe-27(it132); LGV: dpy-11(e224), myo-3(st386), ego-3(om40); and LGX: lon-2(e678). Rearrangements: hDf10, nDf24, nDf25, nDf30, qDf7, qDf8, maDf4, mnDf16, mnDf68, mnDf89, nDf40, nT1(IV;V), nDf41, eDf19, mDf7, sDf23, stDf7, stDf8, ctDf1, and nDf42.
Isolation of ts Stu mutants: Two procedures were used to isolate ts Stu mutants. Both were based on methods described for the isolation of maternal-effect lethal (Mel) mutants (Priess et al. 1987; Kemphues et al.1988b). The basic approach involves the use of a strain carrying a mutation that prevents egg laying, such as sem-2(n1343). These hermaphrodites are still capable of internal self-fertilization, and the embryos that are produced accumulate in the uterus, where they hatch. The trapped larvae feed on maternal tissue and transform the mother into a carcass of writhing larvae bound by an unpalatable cuticle (bag-of-worms phenotype), from which the offspring eventually escape. Mutations that block the production of viable offspring allow sem-2 hermaphrodites to survive. Among the survivors, hermaphrodites homozygous for a maternal-effect lethal mutation can be identified by the presence of a distended uterus full of dead refractile fertilized eggs. In contrast, sterile mutants lack fertilized eggs—but, depending on the severity and type of mutation, they may possess oocytes and sperm (fertilization-defective), lack oocytes and/or sperm (gametogenesis-defective) or lack a normal somatic gonad (gonadogenesis-defective).
Method 1: A population of sem-2 animals was treated with 40 mm ethyl methanesulfonate (EMS) essentially as described by Brenner (1974). Twenty-five to 50 mutagenized animals were picked to individual 60-mm plates and allowed to produce an F1 generation at 20° or 25°. From each culture, 12 F1 hermaphrodites at the L4 larval stage were picked to individual wells of 12-well tissue-culture plates containing NGM media seeded with E. coli OP50. At this point, all animals were incubated at 25°. When a young F3 generation was visible, plates were screened under a dissecting microscope for surviving F2 adults. Wells containing F2 Stu animals were scored as positive. Mutations were recovered from heterozygotes in the F3 population and maintained by picking 8 to 12 animals to individual plates and scoring each for the production of one-quarter Stu progeny. At the same time, lines were tested for temperature sensitivity by individually transferring 6 to 8 gravid F3 animals to 16° and scoring their offspring for the Stu phenotype. Lines that produced Stu animals at low temperature were discarded; those in which Stu animals were not detected were tested again. Lines in which the Stu phenotype was reproducibly absent at low temperature were retained, and an attempt to derive a homozygous line at 16° was made. In all but one case (that of abc-1), we were able to establish a homozygous line. At 25°, abc-1 homozygotes possessed a strong Stu phenotype, while at low temperature they developed into fertile adults that produced only dead eggs. To propagate abc-1, we placed it over the balancer chromosome nT1(IV;V), which confers dominant uncoordinated and recessive lethal phenotypes (Edgley et al. 1995). This balanced stock provided a source of abc-1 homozygotes for most of the work described in this article.
Using Method 1, we screened 3678 F1 animals, or 7356 haploid genomes, for a ts Stu phenotype and obtained the mutations stu-8(oj1), abc-1(oj2), spd-1(oj5), and zyg-1(oj7).
Method 2: A modification of Method 1 allowed us to screen for ts Stu mutants more efficiently. A key feature of Method 2 was that F2 animals carrying dead eggs were first identified at high temperature, then screened for temperature sensitivity by being transferred individually to low temperature. Some of the animals that carried an emb mutation were able to produce a few viable offspring after the temperature decrease. These offspring were able to found lines of homozygous animals that were screened for the Stu phenotype.
The basic approach was as follows: Large quantities of sem-2 worms were cultivated in liquid media at room temperature, essentially as described (Sulston and Hodgkin 1988). The larger quantities were required partly because of the larger number of animals that could be screened and partly because this method employs a synchronization step in which most of the population is killed off. Part of a culture containing many L4 larvae was spun down at 2500 rpm for 5 min, washed several times with M9 buffer (85.6 mm NaCl, 42.3 mm Na2HPO4, 22 mm KH2PO4, 1 mm MgSO4), and suspended in fresh M9 buffer. The worm suspension was adjusted to 40 mm EMS, agitated for 3 hr at room temperature, then allowed to rest undisturbed for an additional hour. Worms that had settled to the bottom of the tube were recovered, transferred to fresh liquid-culture media, and grown with constant agitation at room temperature until many F1 gravid adults were visible. The worms were collected by centrifugation as before, washed several times, and suspended in 7.5 ml M9 buffer. To obtain a semisynchronized population of F2 animals, the volume of the worm suspension was adjusted to 35 ml with a solution of 0.25 m KOH and 1.3% sodium hypochlorite. F1 adults and larvae were dissolved in the basic hypochlorite solution, but unhatched F2 embryos survived. The isolated F2 eggs were distributed at a density between 250 and 750 eggs per 100-mm NGM plate, and the plates were placed at 16° until the majority of animals had reached the L4 stage. At this time, plates were moved to 25°, where they remained until most of the F2 adults had been consumed. Surviving F2 hermaphrodites carrying dead eggs—and presumably homozygous for an emb mutation—were picked individually to 12-well tissue-culture plates. The plates were stored at 16° for 2 to 3 weeks, and each well was scored for the presence of viable F3 animals. Lines were retested for the Emb phenotype by shifting four L4 animals to 25°. Secondary testing revealed many of the lines to be false positives, probably the result of synthetic effects (many independent mutations contributing to the Mel phenotype), single mutations with a variable phenotype, or picking errors. Lines with a reproducible Emb phenotype were tested for the presence of the Stu phenotype. This was accomplished by examining any surviving adult progeny from the secondary Emb test or by shifting carcasses full of young larvae to high temperature. Lines that displayed both the Emb and Stu phenotypes were retained.
The number of haploid genomes screened was estimated as follows: First, the frequency of F1 animals carrying an emb mutation was determined by picking gravid F1 adults to single-culture wells just before the synchronization step. These animals were exposed to the same temperature regime as the isolated F2 eggs. F1 mothers heterozygous for an emb mutation were identified by examining their offspring for individuals carrying dead eggs. We found that under these conditions, approximately one in three F1 hermaphrodites carried an emb mutation. Thus, for each surviving F2 adult picked to low temperature, the equivalent of three F1 animals, or six haploid genomes, were screened. As we tested approximately 7900 F2 animals for temperature sensitivity, we estimate that 47,400 haploid genomes were screened using Method 2. The ts mutations stu-9(oj13), stu-10(oj14), stu-11(oj18), stu-12(oj21), slo-1(oj23), stu-13(oj24), stu-14(oj26), stu-15(oj28), spd-2(oj29), stu-16(oj30), stu-17(oj31), stu-18(oj32), stu-19(oj33), cyk-2(oj34), and spd-3(oj35) were identified with this approach.
It should be stressed that the frequency with which ts Stu mutations were identified using Method 2 probably underestimates the frequency at which they were induced by EMS mutagenesis. Animals homozygous for a ts emb mutation would not have been identified had they failed to produced progeny after the shift to low temperature. This could have occurred if the effect of a ts mutation were irreversible or if the F2 mother had run out of sperm prior to the temperature decrease.
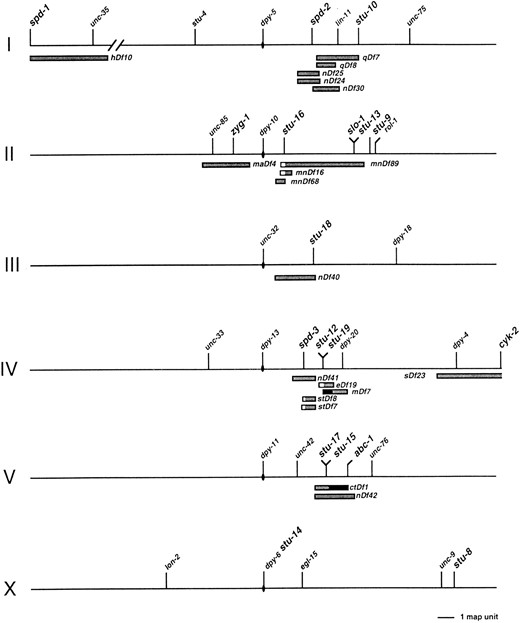
Backcrossing, mapping, and complementation analysis: All mutations were backcrossed at least twice to N2 stocks to remove the sem-2 marker and any extraneous mutations produced during mutagenesis. Selection of backcrossed lines was based on expression of either the Stu or Emb phenotype; in all cases, the unselected phenotype cosegregated, indicating that the two phenotypes were linked. Assignment of mutations to specific chromosomes was performed as described by Brenner (1974), using the following markers: dpy-5 I, rol-1 II, unc-32 III, unc-5 IV, dpy-11 V, and lon-2 X. Despite repeated attempts, we were not able to detect linkages between any of these markers and the stu-11 mutation. We have decided to include stu-11 in the work presented here to provide the most complete description of the screening results. Once a mutation was assigned to a specific chromosome, we determined its position within the linkage group using two- and three-factor mapping techniques (Brenner 1974). In these crosses, Stu mutations were followed by scoring either the sterile phenotype or the Emb phenotype. The map position of each Stu mutation shown in Figure 1 was established by scoring a minimum of 35 recombinant chromosomes. All raw mapping data have been submitted to the C. elegans genomic database AceDB (Eeckman and Durbin 1995).
To confirm the positions obtained with three-factor mapping, Stu mutations were tested for complementation with genetic deficiencies that mapped within the regions of interest. Stu mutations were also tested for allelism with closely linked (0.25–1.0 map units) mutations known to confer a sterile, Stu, lethal, Mel, or Emb phenotype (see Table 1). Noncomplementation was indicated by the presence of sterile or Stu progeny. In cases where the test marker appeared to complement the Stu mutation, we verified the results by confirming the presence of fertile animals carrying both the marker and the Stu mutation.
Penetrance tests: To determine the types and frequencies of postembryonic defects caused by Stu mutations, we exposed mutant animals to the restrictive temperature at the completion of embryogenesis. Plates containing worms grown at 16° were scanned under the high-power lens (×160 total magnification) of a Wild Kombistereo dissecting microscope for unhatched eggs containing threefold embryos, a postmorphogenic state marked by an adultlike body plan, a length three times that of the egg-shell, and extensive writhing activity. Each embryo was placed in one well of a 12-well plate, and the plates were transferred to high temperature for several days. Each well was examined for the ultimate fate of the animal. Some animals arrested during larval development. These were often misshapen and necrotic, making it difficult to assign them to one of the four (L1–L4) larval stages. Thus, we estimated stages on the basis of size alone and assigned animals to one of the following classes: early larval lethal (L1/L2), midlarval lethal (L2/L3), or late larval lethal (L3/L4). Hermaphrodites that developed to adulthood were scored for fertility. The absence of fertilized eggs was scored as sterility regardless of the appearance of the adult: We did not distinguish between the presence or absence of gametes or a normal somatic gonad. Some of the adults were also scored for vulval defects. This was usually accomplished using the high-power lens of the dissecting microscope.
To measure penetrance of the Emb phenotype, homozygous L4 larvae or young adults from each strain were picked individually to 35-mm NGM plates at 25°. Twenty-four hours later, each animal was removed and transferred to a second plate for an additional 24 hr. Dead (unhatched) eggs were counted on both sets of plates 1 day after adults were removed. As embryogenesis is completed in about 14 hr at this temperature (Wood et al. 1980), eggs that remained unhatched after 24 hr were scored as dead. After the dead eggs had been counted, the plates were returned to 25°, and the next day all viable progeny were counted, keeping track of the number of animals that had arrested at the L1 larval stage. While in most instances animals that escaped embryonic lethality developed into sterile adults, for two mutations a significant number of escapers arrested at the L1 stage. These cases are noted in Table 4. For this penetrance (P) test, as well as the rescue and selfing tests described below, a minimum of four hermaphrodites from each line were brooded or crossed.
Parental tests: To determine whether embryonic lethality was caused by failed maternal or zygotic expression of wild-type activity, mutant lines were subject to two additional tests. In the male rescue (R) test (Wood et al. 1980), homozygous mutant hermaphrodites were mated to wild-type males, and the viability of offspring was compared to that of unmated control hermaphrodites (from the P test). A higher survival rate among the progeny of a mated hermaphrodite was interpreted as rescue.
The R test was performed in a manner similar to that of the P test. Each homozygous mutant L4 larva or young adult was placed at 25° in the presence of four N2 males. After 24 hr the adult males were removed, and the hermaphrodite was transferred to a second plate. After an additional 24 hr at 25°, the hermaphrodite was moved to a third plate. The first and third plates were placed at 16° immediately after the animals were transferred. These were used to assess successful mating. The second plate was left at 25° for 24 hr. The viability rate was calculated from the number of dead eggs and live progeny present on the second plate. To confirm that mating had taken place, all three plates were examined for the presence of progeny males. For several strains, mating could not be confirmed in this manner, as mutant hermaphrodites did not produce live progeny. In these cases, the test was performed using plg-1 males, which deposit a gelatinous plug over the vulva during copulation. Mating was confirmed by scoring for the presence of a plug at the time males were removed.
To determine whether maternal expression of wild-type gene activity could suffice for embryogenesis, we employed the selfing (S) test (Wood et al. 1980). A hermaphrodite heterozygous for a Stu mutation was allowed to self-fertilize at the restrictive temperature, and the viability of offspring was determined as described for the P test. Heterozygotes were obtained by mating N2 males with morphologically marked homozygous Stu mutant stocks and picking nonmarked offspring. The rate of survival among the homozygous mutant offspring indicated the degree of maternal rescue.
DNA staining: Animals grown at 25° were washed off seeded plates with M9 buffer and transferred to a microfuge tube. The animals were washed several times with M9 buffer and fixed in 100% methanol for 10 min at room temperature. Worms were removed from the fixative by centrifugation and stained in a solution of 0.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 10 min at room temperature. The specimens were mounted on a microscope slide, and images were obtained under epifluorescence illumination using a Photometrics (Tucson, AZ) SenSys KAF 1400 CCD camera.
Multiple focal plane time-lapse imaging: Early development of mutant embryos was analyzed with a system capable of making time-lapse differential interference contrast (DIC) images of multiple focal planes (Thomas et al. 1996). The system utilized for these studies contained a Nikon (Melville, NY) Optiphot-2 microscope mounted with a Hamamatsu (Bridgewater, NJ) C2400 video camera. The video signal from the camera was fed to an AG-5 frame-grabber card (Scion Corporation, Frederick, MD) installed in a Macintosh PowerPC 7600 computer. The AG-5card supported both frame averaging and contrast enhancement. Automated focus control was obtained using a stage motor driven by a Ludl MAC 2000 controller. The controller was connected to the computer via the serial port. The four-dimensional (4D) image acquisition, data translation, and viewing and analysis applications used in these studies were written by Charles Thomas of the Integrated Microscopy Resource (IMR), University of Wisconsin. All software is available free of charge at the IMR Website, http://www.bocklabs.wisc.edu/imr/home.htm/.
To prepare embryos for 4D analysis, mutant animals at the L4 larval stage or older were shifted to 25° for approximately 24 to 36 hr. Young embryos were quickly dissected from gravid mothers in egg salts (118 mm NaCl, 48 mm KCl, 2 mm CaCl2, 2 mm MgCl2, and 5 mm HEPES, pH 7.4) and transferred by mouth pipet onto a cushion of solidified 3% agarose in egg salts that had been cast on a glass slide. A coverslip was placed gently over the drop, the edges were sealed with molten vaseline, and the slide was transferred to the microscope. All recordings were performed at 25°. Temperature was controlled via the room thermostat or locally, using a hair dryer equipped with a feedback thermocouple to heat the microscope stage.
RESULTS
Identification of a novel set of conditional cell-division mutations: We have employed a new approach to identifying conditional cell-division mutations in C. elegans that is based on expression of two phenotypes known to be associated with cell-division failure. Nineteen EMS-generated ts mutations that conferred both Emb and Stu phenotypes were identified. Eighteen of these mutations were positioned on the genetic map using two- and three-factor mapping techniques (materials and methods). Where possible, the positions were confirmed using genetic deficiencies that mapped within the appropriate regions. The mutations mapped to all five autosomes and the X chromosome (Figure 1). Closely linked Stu mutations were tested for allelism, and in all cases the mutations were found to complement one another. Thus, these 18 mutations define 18 distinct genes.
To determine whether any of these 18 Stu mutations represented new alleles of previously identified genes, complementation tests were performed between Stu mutations and any closely linked mutation known to possess a sterile, Stu, lethal, or Emb phenotype. The
Positions of Stu mutations in a simplified genetic map of C. elegans. Each holocentric chromosome is depicted as a horizontal line, with the designated center indicated by a solid oval. The positions of selected genes are indicated by vertical lines; those identified in this screen are shown in a larger font. The mutation oj7 was only partially mapped before it was determined to be an allele of the zyg-1 gene. Thus, the previously established map position of zyg-1 is shown. The positions and extents of chromosomal deficiencies are indicated by the shaded bars beneath the corresponding chromosomes. In some cases, a deficiency appeared to overlap the position of a Stu mutation that it complemented; the unshaded area indicates the extent of overlap. In other cases, a deficiency appeared not to overlap the position of a Stu mutation that it failed to complement; the distance between the deficiency breakpoint and the Stu mutation is indicated by a solid bar.
results of these complementation tests are summarized in Table 1. Despite exhaustive testing, in only one case did we find that a Stu mutation failed to complement a known mutation. The mutation oj7 failed to complement zyg-1(b1) (Table 1). We conclude that the new approach is capable of identifying many new genes.
Stu mutations affect many aspects of postembryonic development: The lin-5 and lin-6 mutations block postembryonic cell division, leading to a Stu phenotype (Albertson et al. 1978; Horvitz and Sulston 1980; Sulston and Horvitz 1981). To estimate to what extent these new mutations might affect the postembryonic cell divisions, we exposed mutant animals to the restrictive temperature for the entirety of postembryonic development and scored for viability, fertility, and vulval morphology.
Complementation analysis
| Mutationa . | Complemented by . | Not complemented by . |
|---|---|---|
| stu-8(oj1) X | ||
| abc-1(oj2) V | stu-15(oj28), ego-3(om40) | nDf42, ctDf1 |
| spd1(oj5) I | mex-3(zu155), spe-8(hc40) | hDf10 |
| zyg-1(oj7) II | lin-5(e1348) | maDf4, zyg-1(b1) |
| stu-9(oj13) II | mnDf89, slo-1(oj23), stu-13(oj24) | |
| stu-10(oj14) I | nDf25, nDf24, qDf8 | qDf7 |
| stu-11(oj18) | ||
| stu-12(oj21) IV | nDf41, stu-19(oj33), emb-3(hc59), mes-6(bn66), let-296(s1250), let-292(s1146), let-297(s1989) | |
| slo-1(oj23) II | stu-13(oj24), stu-9(oj13), mel-22(it30), mel-15(it38), let-266(mn194), let-251(mn95) | mnDf89 |
| stu-13(oj24) II | slo-1(of23), stu-9(of13), mel-22(it30), mel-15(it38), let-266(mn194), let-251(mn95) | mnDf89 |
| stu-14(oj26) X | ||
| stu-15(oj28) V | stu-17(oj31), abc-1(oj2), myo-3(st386) | nDf42, ctDf1 |
| spd-2(oj29) I | nDf30, fer-1(hc13), Jet-541(h886), Jet-536(h882) | nDf25, nDf24 |
| stu-16(oj30) II | mnDf16, mnDf89, mel-11(it26), mel-9(b293), let-240(mn209), let-31(mn31), let-239(mn93), let-23(sy10), let-252(mn100), let-19(mn19), let-238 (mn229), evl-20(ar103) | mnDf68 |
| stu-17(oj31) V | stu-15(oj28), myo-3(st386) | ctDf1 |
| stu-18(oj32) III | ooc-4(e2078), cul-1(e1756) | nDf40 |
| stu-19(oj33) IV | nDf41, eDf19, stu-12(oj21), emb-3(hc59), mes-6(bn66), mel-24(ct59), let-54(s44), let-71(s692), let-64(s216), let-73(s685), let-655(s1748) gon-1(q518), gon-4(e2575) | mDf7 |
| cyk-2(oj34) IV | sDf23 | |
| spd-3(oj35) IV | stDf8, stDf7, eDf19, emb-26(g47), emb-31(g55), evl-7(ar108), fem-1(e1991), spe-27(it132) | nDf41 |
| Mutationa . | Complemented by . | Not complemented by . |
|---|---|---|
| stu-8(oj1) X | ||
| abc-1(oj2) V | stu-15(oj28), ego-3(om40) | nDf42, ctDf1 |
| spd1(oj5) I | mex-3(zu155), spe-8(hc40) | hDf10 |
| zyg-1(oj7) II | lin-5(e1348) | maDf4, zyg-1(b1) |
| stu-9(oj13) II | mnDf89, slo-1(oj23), stu-13(oj24) | |
| stu-10(oj14) I | nDf25, nDf24, qDf8 | qDf7 |
| stu-11(oj18) | ||
| stu-12(oj21) IV | nDf41, stu-19(oj33), emb-3(hc59), mes-6(bn66), let-296(s1250), let-292(s1146), let-297(s1989) | |
| slo-1(oj23) II | stu-13(oj24), stu-9(oj13), mel-22(it30), mel-15(it38), let-266(mn194), let-251(mn95) | mnDf89 |
| stu-13(oj24) II | slo-1(of23), stu-9(of13), mel-22(it30), mel-15(it38), let-266(mn194), let-251(mn95) | mnDf89 |
| stu-14(oj26) X | ||
| stu-15(oj28) V | stu-17(oj31), abc-1(oj2), myo-3(st386) | nDf42, ctDf1 |
| spd-2(oj29) I | nDf30, fer-1(hc13), Jet-541(h886), Jet-536(h882) | nDf25, nDf24 |
| stu-16(oj30) II | mnDf16, mnDf89, mel-11(it26), mel-9(b293), let-240(mn209), let-31(mn31), let-239(mn93), let-23(sy10), let-252(mn100), let-19(mn19), let-238 (mn229), evl-20(ar103) | mnDf68 |
| stu-17(oj31) V | stu-15(oj28), myo-3(st386) | ctDf1 |
| stu-18(oj32) III | ooc-4(e2078), cul-1(e1756) | nDf40 |
| stu-19(oj33) IV | nDf41, eDf19, stu-12(oj21), emb-3(hc59), mes-6(bn66), mel-24(ct59), let-54(s44), let-71(s692), let-64(s216), let-73(s685), let-655(s1748) gon-1(q518), gon-4(e2575) | mDf7 |
| cyk-2(oj34) IV | sDf23 | |
| spd-3(oj35) IV | stDf8, stDf7, eDf19, emb-26(g47), emb-31(g55), evl-7(ar108), fem-1(e1991), spe-27(it132) | nDf41 |
Gene names are as follows: stu, sterile uncoordinated; abc, anaphase bridging of chromatin; spd, spindle defective; zyg, zygote defective; slo, slow development; cyk, cytokinesis defective.
Complementation analysis
| Mutationa . | Complemented by . | Not complemented by . |
|---|---|---|
| stu-8(oj1) X | ||
| abc-1(oj2) V | stu-15(oj28), ego-3(om40) | nDf42, ctDf1 |
| spd1(oj5) I | mex-3(zu155), spe-8(hc40) | hDf10 |
| zyg-1(oj7) II | lin-5(e1348) | maDf4, zyg-1(b1) |
| stu-9(oj13) II | mnDf89, slo-1(oj23), stu-13(oj24) | |
| stu-10(oj14) I | nDf25, nDf24, qDf8 | qDf7 |
| stu-11(oj18) | ||
| stu-12(oj21) IV | nDf41, stu-19(oj33), emb-3(hc59), mes-6(bn66), let-296(s1250), let-292(s1146), let-297(s1989) | |
| slo-1(oj23) II | stu-13(oj24), stu-9(oj13), mel-22(it30), mel-15(it38), let-266(mn194), let-251(mn95) | mnDf89 |
| stu-13(oj24) II | slo-1(of23), stu-9(of13), mel-22(it30), mel-15(it38), let-266(mn194), let-251(mn95) | mnDf89 |
| stu-14(oj26) X | ||
| stu-15(oj28) V | stu-17(oj31), abc-1(oj2), myo-3(st386) | nDf42, ctDf1 |
| spd-2(oj29) I | nDf30, fer-1(hc13), Jet-541(h886), Jet-536(h882) | nDf25, nDf24 |
| stu-16(oj30) II | mnDf16, mnDf89, mel-11(it26), mel-9(b293), let-240(mn209), let-31(mn31), let-239(mn93), let-23(sy10), let-252(mn100), let-19(mn19), let-238 (mn229), evl-20(ar103) | mnDf68 |
| stu-17(oj31) V | stu-15(oj28), myo-3(st386) | ctDf1 |
| stu-18(oj32) III | ooc-4(e2078), cul-1(e1756) | nDf40 |
| stu-19(oj33) IV | nDf41, eDf19, stu-12(oj21), emb-3(hc59), mes-6(bn66), mel-24(ct59), let-54(s44), let-71(s692), let-64(s216), let-73(s685), let-655(s1748) gon-1(q518), gon-4(e2575) | mDf7 |
| cyk-2(oj34) IV | sDf23 | |
| spd-3(oj35) IV | stDf8, stDf7, eDf19, emb-26(g47), emb-31(g55), evl-7(ar108), fem-1(e1991), spe-27(it132) | nDf41 |
| Mutationa . | Complemented by . | Not complemented by . |
|---|---|---|
| stu-8(oj1) X | ||
| abc-1(oj2) V | stu-15(oj28), ego-3(om40) | nDf42, ctDf1 |
| spd1(oj5) I | mex-3(zu155), spe-8(hc40) | hDf10 |
| zyg-1(oj7) II | lin-5(e1348) | maDf4, zyg-1(b1) |
| stu-9(oj13) II | mnDf89, slo-1(oj23), stu-13(oj24) | |
| stu-10(oj14) I | nDf25, nDf24, qDf8 | qDf7 |
| stu-11(oj18) | ||
| stu-12(oj21) IV | nDf41, stu-19(oj33), emb-3(hc59), mes-6(bn66), let-296(s1250), let-292(s1146), let-297(s1989) | |
| slo-1(oj23) II | stu-13(oj24), stu-9(oj13), mel-22(it30), mel-15(it38), let-266(mn194), let-251(mn95) | mnDf89 |
| stu-13(oj24) II | slo-1(of23), stu-9(of13), mel-22(it30), mel-15(it38), let-266(mn194), let-251(mn95) | mnDf89 |
| stu-14(oj26) X | ||
| stu-15(oj28) V | stu-17(oj31), abc-1(oj2), myo-3(st386) | nDf42, ctDf1 |
| spd-2(oj29) I | nDf30, fer-1(hc13), Jet-541(h886), Jet-536(h882) | nDf25, nDf24 |
| stu-16(oj30) II | mnDf16, mnDf89, mel-11(it26), mel-9(b293), let-240(mn209), let-31(mn31), let-239(mn93), let-23(sy10), let-252(mn100), let-19(mn19), let-238 (mn229), evl-20(ar103) | mnDf68 |
| stu-17(oj31) V | stu-15(oj28), myo-3(st386) | ctDf1 |
| stu-18(oj32) III | ooc-4(e2078), cul-1(e1756) | nDf40 |
| stu-19(oj33) IV | nDf41, eDf19, stu-12(oj21), emb-3(hc59), mes-6(bn66), mel-24(ct59), let-54(s44), let-71(s692), let-64(s216), let-73(s685), let-655(s1748) gon-1(q518), gon-4(e2575) | mDf7 |
| cyk-2(oj34) IV | sDf23 | |
| spd-3(oj35) IV | stDf8, stDf7, eDf19, emb-26(g47), emb-31(g55), evl-7(ar108), fem-1(e1991), spe-27(it132) | nDf41 |
Gene names are as follows: stu, sterile uncoordinated; abc, anaphase bridging of chromatin; spd, spindle defective; zyg, zygote defective; slo, slow development; cyk, cytokinesis defective.
Nearly all mutations conferred some degree of larval lethality (Tables 2 and 3). In particular, abc-1, spd-2, and stu-18 animals exhibited high levels of larval lethality, suggesting that these genes might be required postembryonically for viability. However, for spd-2 and stu-18, we found that similar percentages of animals arrested as larvae at 16° and 25°—indicating that, in these cases, larval lethality occurred independent of the temperature shift. It is possible that at 16°, many spd-2 and stu-18 animals experience a low level of random cell-division failure; some might be healthy enough to hatch but too sick to develop beyond the larval stages.
For 10 mutant lines, the penetrance of the sterile phenotype was nearly complete: all or almost all of the animals that developed to adulthood were sterile. Among this group of mutants were those with the most severe defects. stu-8, abc-1, zyg-1, stu-15, and spd-2 animals were extremely uncoordinated, lacked a normal gonad, and possessed highly penetrant vulval defects (Tables 2 and 3). Adult stu-9, stu-10, stu-19, and cyk-2 hermaphrodites were also uniformly sterile, but the gonad appeared to be well developed, and the Unc phenotype tended to be weak and variable. For spd-3, the sterile phenotype was nearly absolute: only one of 38 adults scored was fertile.
This group of mutations strongly affected another aspect of postembryonic development that requires cell division: vulval development (Table 2). In general, the penetrance of vulval defects correlated with the severity of the Stu phenotype. All zyg-1 animals failed to form a normal vulva; most were scored as vulvaless (Vul), as they lacked a recognizable structure altogether. A few contained an apparently nonfunctional vulva that protruded from the body (Pvl), and one possessed multiple vulvae (Muv). Likewise, the other lines with a strong Stu phenotype, stu-8, abc-1, stu-15, and spd-2, contained few animals with a normal vulva. Among these animals the most common defect was a Pvl phenotype. The mutations stu-9, stu-10, stu-19, and cyk-2 conferred milder Stu phenotypes, and accordingly the vulval defects were less penetrant. The only exception to the strong positive correlation between the Stu and vulva phenotypes was spd-3. These animals possessed a somewhat mild Stu phenotype capable of nearly normal movement but exhibited a uniform Vul phenotype.
The remaining nine lines, spd-1, stu-11, stu-12, slo-1,
Postembryonic phenotypes caused by Stu mutations
| . | Larval arresta . | Adult Phenotypeb . | Vulval morphologyc . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation . | Early . | Mid . | Late . | Sterile . | Fertile . | Male . | Normal . | Pv1 . | Vu1 . | Muv . |
| stu-8(oj1) | 4 | 0 | 0 | 47 | 0 | 0 | 1 | 17 | 2 | 0 |
| abc-1(oj2)d | 9 | 0 | 1 | 32 | 0 | 0 | 1 | 26 | 3 | 0 |
| spd-1(oj5) | 6 | 0 | 0 | 57 | 12 | 0 | 0 | 68 | 0 | 0 |
| zyg-1(oj7) | 5 | 0 | 0 | 29 | 0 | 0 | 0 | 2 | 25 | 1 |
| stu-9(oj13) | 1 | 0 | 1 | 65 | 0 | 0 | 38 | 0 | 0 | 0 |
| stu-10(oj14) | 1 | 0 | 0 | 46 | 0 | 10 | 27 | 19 | 0 | 0 |
| stu-11(oj18)e | 0 | 0 | 0 | 6 | 77 | 0 | 24 | 0 | 0 | 0 |
| stu-12(oj21)e | 3 | 0 | 0 | 13 | 73 | 1 | 24 | 2 | 0 | 0 |
| slo-1(oj23) | 2 | 0 | 0 | 46 | 14 | 4 | 37 | 0 | 0 | 0 |
| stu-13(oj24)e | 1 | 0 | 0 | 3 | 86 | 1 | 23 | 0 | 0 | 0 |
| stu-14(oj26) | 4 | 0 | 0 | 26 | 7 | 0 | 29 | 1 | 0 | 0 |
| stu-15(oj28) | 1 | 0 | 0 | 65 | 0 | 0 | 2 | 40 | 3 | 4 |
| spd-2(oj29) | 34 | 0 | 0 | 25 | 0 | 1 | 1 | 8 | 14 | 0 |
| stu-16(oj30) | 1 | 0 | 1 | 38 | 23 | 0 | 35 | 16 | 1 | 0 |
| stu-17(oj31) | 0 | 0 | 0 | 27 | 11 | 1 | 10 | 0 | 22 | 0 |
| stu-18(oj32)e | 9 | 3 | 4 | 19 | 24 | 0 | 18 | 0 | 0 | 1 |
| stu-19(oj33) | 1 | 0 | 1 | 42 | 0 | 0 | 42 | 0 | 0 | 0 |
| cyk-2(oj34) | 3 | 0 | 0 | 52 | 0 | 0 | 27 | 0 | 0 | 0 |
| spd-3(oj35) | 2 | 0 | 0 | 37 | 1 | 0 | 0 | 0 | 19 | 0 |
| . | Larval arresta . | Adult Phenotypeb . | Vulval morphologyc . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation . | Early . | Mid . | Late . | Sterile . | Fertile . | Male . | Normal . | Pv1 . | Vu1 . | Muv . |
| stu-8(oj1) | 4 | 0 | 0 | 47 | 0 | 0 | 1 | 17 | 2 | 0 |
| abc-1(oj2)d | 9 | 0 | 1 | 32 | 0 | 0 | 1 | 26 | 3 | 0 |
| spd-1(oj5) | 6 | 0 | 0 | 57 | 12 | 0 | 0 | 68 | 0 | 0 |
| zyg-1(oj7) | 5 | 0 | 0 | 29 | 0 | 0 | 0 | 2 | 25 | 1 |
| stu-9(oj13) | 1 | 0 | 1 | 65 | 0 | 0 | 38 | 0 | 0 | 0 |
| stu-10(oj14) | 1 | 0 | 0 | 46 | 0 | 10 | 27 | 19 | 0 | 0 |
| stu-11(oj18)e | 0 | 0 | 0 | 6 | 77 | 0 | 24 | 0 | 0 | 0 |
| stu-12(oj21)e | 3 | 0 | 0 | 13 | 73 | 1 | 24 | 2 | 0 | 0 |
| slo-1(oj23) | 2 | 0 | 0 | 46 | 14 | 4 | 37 | 0 | 0 | 0 |
| stu-13(oj24)e | 1 | 0 | 0 | 3 | 86 | 1 | 23 | 0 | 0 | 0 |
| stu-14(oj26) | 4 | 0 | 0 | 26 | 7 | 0 | 29 | 1 | 0 | 0 |
| stu-15(oj28) | 1 | 0 | 0 | 65 | 0 | 0 | 2 | 40 | 3 | 4 |
| spd-2(oj29) | 34 | 0 | 0 | 25 | 0 | 1 | 1 | 8 | 14 | 0 |
| stu-16(oj30) | 1 | 0 | 1 | 38 | 23 | 0 | 35 | 16 | 1 | 0 |
| stu-17(oj31) | 0 | 0 | 0 | 27 | 11 | 1 | 10 | 0 | 22 | 0 |
| stu-18(oj32)e | 9 | 3 | 4 | 19 | 24 | 0 | 18 | 0 | 0 | 1 |
| stu-19(oj33) | 1 | 0 | 1 | 42 | 0 | 0 | 42 | 0 | 0 | 0 |
| cyk-2(oj34) | 3 | 0 | 0 | 52 | 0 | 0 | 27 | 0 | 0 | 0 |
| spd-3(oj35) | 2 | 0 | 0 | 37 | 1 | 0 | 0 | 0 | 19 | 0 |
Number of animals that arrested during the L1/L2 (early), L2/L3 (middle), or L3/L4 (late) larval stages.
Number of animals that developed to adulthood and were determined to be sterile hermaphrodites, fertile hermaphrodites, or males. The total number of animals scored is the sum of the number of animals exhibiting larval arrest and the number of animals scored for adult fates.
A subset of adult hermaphrodites were scored for vulval morphology. Shown are the number of animals with one of the following morphologies: normal, not appreciably dissimilar from wild type; Pvl, protuding vulva; Vul, vulva absent; Muv, multiple (typically abnormal) value.
Test performed using abc-1 heterozygous parents. Offspring of abc-1(oj2)/ + parents were transferred at the three fold stage of embryogenesis to 25°. Of 144 animals, 102 were phenotypically wild type. The remainder, which arrested as larvae or developed into sterile adults as indicated in the table, were inferred to be abc-1 homozygotes. Penetrance of the sterile phenotype was confirmed in independent tests using the abc-1/nT1 strain, in which abc-1 homozygotes could be unambiguously identified.
In additional tests, these homozygotes were expossed to the restrictive temperature earlier in development by allowing gravid mothers to lay eggs for 3.25 hr at 25°. Of the animals produced during this period, 132 of 133 stu-11 hermaphrodites, 35 of 84 stu-12 hermaphrodites, 58 of 77 stu-13 hermaphrodites, and 51 of 60 stu-18 hermaphrodites developed into sterile adults.
Postembryonic phenotypes caused by Stu mutations
| . | Larval arresta . | Adult Phenotypeb . | Vulval morphologyc . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation . | Early . | Mid . | Late . | Sterile . | Fertile . | Male . | Normal . | Pv1 . | Vu1 . | Muv . |
| stu-8(oj1) | 4 | 0 | 0 | 47 | 0 | 0 | 1 | 17 | 2 | 0 |
| abc-1(oj2)d | 9 | 0 | 1 | 32 | 0 | 0 | 1 | 26 | 3 | 0 |
| spd-1(oj5) | 6 | 0 | 0 | 57 | 12 | 0 | 0 | 68 | 0 | 0 |
| zyg-1(oj7) | 5 | 0 | 0 | 29 | 0 | 0 | 0 | 2 | 25 | 1 |
| stu-9(oj13) | 1 | 0 | 1 | 65 | 0 | 0 | 38 | 0 | 0 | 0 |
| stu-10(oj14) | 1 | 0 | 0 | 46 | 0 | 10 | 27 | 19 | 0 | 0 |
| stu-11(oj18)e | 0 | 0 | 0 | 6 | 77 | 0 | 24 | 0 | 0 | 0 |
| stu-12(oj21)e | 3 | 0 | 0 | 13 | 73 | 1 | 24 | 2 | 0 | 0 |
| slo-1(oj23) | 2 | 0 | 0 | 46 | 14 | 4 | 37 | 0 | 0 | 0 |
| stu-13(oj24)e | 1 | 0 | 0 | 3 | 86 | 1 | 23 | 0 | 0 | 0 |
| stu-14(oj26) | 4 | 0 | 0 | 26 | 7 | 0 | 29 | 1 | 0 | 0 |
| stu-15(oj28) | 1 | 0 | 0 | 65 | 0 | 0 | 2 | 40 | 3 | 4 |
| spd-2(oj29) | 34 | 0 | 0 | 25 | 0 | 1 | 1 | 8 | 14 | 0 |
| stu-16(oj30) | 1 | 0 | 1 | 38 | 23 | 0 | 35 | 16 | 1 | 0 |
| stu-17(oj31) | 0 | 0 | 0 | 27 | 11 | 1 | 10 | 0 | 22 | 0 |
| stu-18(oj32)e | 9 | 3 | 4 | 19 | 24 | 0 | 18 | 0 | 0 | 1 |
| stu-19(oj33) | 1 | 0 | 1 | 42 | 0 | 0 | 42 | 0 | 0 | 0 |
| cyk-2(oj34) | 3 | 0 | 0 | 52 | 0 | 0 | 27 | 0 | 0 | 0 |
| spd-3(oj35) | 2 | 0 | 0 | 37 | 1 | 0 | 0 | 0 | 19 | 0 |
| . | Larval arresta . | Adult Phenotypeb . | Vulval morphologyc . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation . | Early . | Mid . | Late . | Sterile . | Fertile . | Male . | Normal . | Pv1 . | Vu1 . | Muv . |
| stu-8(oj1) | 4 | 0 | 0 | 47 | 0 | 0 | 1 | 17 | 2 | 0 |
| abc-1(oj2)d | 9 | 0 | 1 | 32 | 0 | 0 | 1 | 26 | 3 | 0 |
| spd-1(oj5) | 6 | 0 | 0 | 57 | 12 | 0 | 0 | 68 | 0 | 0 |
| zyg-1(oj7) | 5 | 0 | 0 | 29 | 0 | 0 | 0 | 2 | 25 | 1 |
| stu-9(oj13) | 1 | 0 | 1 | 65 | 0 | 0 | 38 | 0 | 0 | 0 |
| stu-10(oj14) | 1 | 0 | 0 | 46 | 0 | 10 | 27 | 19 | 0 | 0 |
| stu-11(oj18)e | 0 | 0 | 0 | 6 | 77 | 0 | 24 | 0 | 0 | 0 |
| stu-12(oj21)e | 3 | 0 | 0 | 13 | 73 | 1 | 24 | 2 | 0 | 0 |
| slo-1(oj23) | 2 | 0 | 0 | 46 | 14 | 4 | 37 | 0 | 0 | 0 |
| stu-13(oj24)e | 1 | 0 | 0 | 3 | 86 | 1 | 23 | 0 | 0 | 0 |
| stu-14(oj26) | 4 | 0 | 0 | 26 | 7 | 0 | 29 | 1 | 0 | 0 |
| stu-15(oj28) | 1 | 0 | 0 | 65 | 0 | 0 | 2 | 40 | 3 | 4 |
| spd-2(oj29) | 34 | 0 | 0 | 25 | 0 | 1 | 1 | 8 | 14 | 0 |
| stu-16(oj30) | 1 | 0 | 1 | 38 | 23 | 0 | 35 | 16 | 1 | 0 |
| stu-17(oj31) | 0 | 0 | 0 | 27 | 11 | 1 | 10 | 0 | 22 | 0 |
| stu-18(oj32)e | 9 | 3 | 4 | 19 | 24 | 0 | 18 | 0 | 0 | 1 |
| stu-19(oj33) | 1 | 0 | 1 | 42 | 0 | 0 | 42 | 0 | 0 | 0 |
| cyk-2(oj34) | 3 | 0 | 0 | 52 | 0 | 0 | 27 | 0 | 0 | 0 |
| spd-3(oj35) | 2 | 0 | 0 | 37 | 1 | 0 | 0 | 0 | 19 | 0 |
Number of animals that arrested during the L1/L2 (early), L2/L3 (middle), or L3/L4 (late) larval stages.
Number of animals that developed to adulthood and were determined to be sterile hermaphrodites, fertile hermaphrodites, or males. The total number of animals scored is the sum of the number of animals exhibiting larval arrest and the number of animals scored for adult fates.
A subset of adult hermaphrodites were scored for vulval morphology. Shown are the number of animals with one of the following morphologies: normal, not appreciably dissimilar from wild type; Pvl, protuding vulva; Vul, vulva absent; Muv, multiple (typically abnormal) value.
Test performed using abc-1 heterozygous parents. Offspring of abc-1(oj2)/ + parents were transferred at the three fold stage of embryogenesis to 25°. Of 144 animals, 102 were phenotypically wild type. The remainder, which arrested as larvae or developed into sterile adults as indicated in the table, were inferred to be abc-1 homozygotes. Penetrance of the sterile phenotype was confirmed in independent tests using the abc-1/nT1 strain, in which abc-1 homozygotes could be unambiguously identified.
In additional tests, these homozygotes were expossed to the restrictive temperature earlier in development by allowing gravid mothers to lay eggs for 3.25 hr at 25°. Of the animals produced during this period, 132 of 133 stu-11 hermaphrodites, 35 of 84 stu-12 hermaphrodites, 58 of 77 stu-13 hermaphrodites, and 51 of 60 stu-18 hermaphrodites developed into sterile adults.
stu-13, stu-14, stu-16, stu-17, and stu-18, exhibited a lower penetrance of the sterile phenotype. Among these mutants, spd-1 and stu-16 animals exhibited a number of very strong defects. Most spd-1 adults were thin, lacked a functional gonad, and exhibited very poor mobility. Twelve of the 69 spd-1 adults scored were able to produce a few dead embryos. However, these fertile spd-1 animals maintained most of the characteristics of their sterile siblings: they were thin, incapable of normal movement, and possessed vulval defects identical to those of sterile siblings. This suggests that the gonadal lineages are less sensitive to the spd-1 mutation than are the vulval or neuronal lineages. Many stu-16 animals also exhibited striking defects in motility and development of the vulva and gonad. Like spd-1, the Unc phenotype of stu-16 appeared more penetrant than the sterile phenotype (data not shown).
Despite being isolated on the basis of a nearly complete Stu phenotype, four of the mutant lines exhibited low penetrance of this phenotype under the test conditions. A majority of stu-11, stu-12, stu-13, and stu-18 animals developed into fertile adults. One possible explanation for this discrepancy was the fact that the animals were shifted to the restrictive temperature earlier in development during the screen than during these tests. To test this possibility, we again shifted stu-11, stu-12, stu-13, and stu-18 animals to 25° earlier in development. Under these conditions, all four lines exhibited a higher penetrance of the sterile phenotype. Nearly all stu-11 and stu-18 animals and most stu-13 animals developed into sterile adults (see footnote to Table 2). These results indicate that, in these cases, either the sterility results from defects in early embryogenesis or an earlier shift is required to sufficiently reduce the amount of wild-type gene activity prior to postembryonic development.
Summary of mutant phenotypes
| Mutation . | Cytological phenotype . | Emb phenotypea . | Larval arrestb . | Sterile phenotype(% penetrance) . | Unc phenotype (severity) . | Vulval defectsc . |
|---|---|---|---|---|---|---|
| stu-8(oj1) | Moderate (mm) | + | 100 | Strong | P, V | |
| abc-1(oj2) | Nuclear division defective | Strong (mm) | +++ | 100 | Strong | P, V |
| spd-1(oj5) | Spindle defective/cytokinesis defective | Strong (mm) | + | >100 | Strong | P |
| zyg-1(oj7) | Centrosome duplication defective | Strong (mm) | ++ | 100 | Strong | V, P, M |
| stu-9(oj13) | Moderate (mm) | + | 100 | Weak | ||
| stu-10(oj14) | Abnormal spindle alignment | Moderate (mm) | + | 100 | Weak | P |
| stu-11(oj18) | Abnormal spindle alignment | Moderate (mm) | >100 | Weak | ||
| stu-12(oj21) | Moderate (mm) | + | >100 | Weak | P | |
| slo-1(oj23) | Slow development | Strong (mn) | + | >100 | Weak | |
| stu-13(oj24) | Strong (mn) | + | >100 | Weak | ||
| stu-14(oj26) | Moderate | ++ | >100 | Weak | P | |
| (mm) | ||||||
| stu-15(oj28) | Strong (mn) | + | 100 | Strong | P, M, V | |
| spd-2(oj29) | Abnormal microtubule organization | Strong (mm) | +++ | 100 | Strong | V, P |
| stu-16(oj30) | Moderate (mn) | + | >100 | Strong | P, V | |
| stu-17(oj31) | Moderate (mm) | >100 | Weak | V | ||
| stu-18(oj32) | Abnormal centrosome morphology | Moderate (mn) | +++ | >100 | Weak | M |
| stu-19(oj33) | Strong (mm) | + | 100 | Weak | ||
| cyk-2(oj34) | Cytokinesis defective | Moderate (mn) | + | 100 | Weak | |
| spd-3(oj35) | Abnormal microtubule organization | Strong (mm) | + | >100 | Weak | V |
| Mutation . | Cytological phenotype . | Emb phenotypea . | Larval arrestb . | Sterile phenotype(% penetrance) . | Unc phenotype (severity) . | Vulval defectsc . |
|---|---|---|---|---|---|---|
| stu-8(oj1) | Moderate (mm) | + | 100 | Strong | P, V | |
| abc-1(oj2) | Nuclear division defective | Strong (mm) | +++ | 100 | Strong | P, V |
| spd-1(oj5) | Spindle defective/cytokinesis defective | Strong (mm) | + | >100 | Strong | P |
| zyg-1(oj7) | Centrosome duplication defective | Strong (mm) | ++ | 100 | Strong | V, P, M |
| stu-9(oj13) | Moderate (mm) | + | 100 | Weak | ||
| stu-10(oj14) | Abnormal spindle alignment | Moderate (mm) | + | 100 | Weak | P |
| stu-11(oj18) | Abnormal spindle alignment | Moderate (mm) | >100 | Weak | ||
| stu-12(oj21) | Moderate (mm) | + | >100 | Weak | P | |
| slo-1(oj23) | Slow development | Strong (mn) | + | >100 | Weak | |
| stu-13(oj24) | Strong (mn) | + | >100 | Weak | ||
| stu-14(oj26) | Moderate | ++ | >100 | Weak | P | |
| (mm) | ||||||
| stu-15(oj28) | Strong (mn) | + | 100 | Strong | P, M, V | |
| spd-2(oj29) | Abnormal microtubule organization | Strong (mm) | +++ | 100 | Strong | V, P |
| stu-16(oj30) | Moderate (mn) | + | >100 | Strong | P, V | |
| stu-17(oj31) | Moderate (mm) | >100 | Weak | V | ||
| stu-18(oj32) | Abnormal centrosome morphology | Moderate (mn) | +++ | >100 | Weak | M |
| stu-19(oj33) | Strong (mm) | + | 100 | Weak | ||
| cyk-2(oj34) | Cytokinesis defective | Moderate (mn) | + | 100 | Weak | |
| spd-3(oj35) | Abnormal microtubule organization | Strong (mm) | + | >100 | Weak | V |
Letters in parentheses indicate genes in which maternal gene expression is required for viability (mm) and genes in which maternal or zygotic expression is sufficient for viability (mn).
Penetrance of larval arrest indicated as follows: +, 0-10%; + +, 10-20%; + + +, >20%.
Vulval defects are listed in order of highest penetrance. P, Pvl; V, Vul; M, Muv.
Summary of mutant phenotypes
| Mutation . | Cytological phenotype . | Emb phenotypea . | Larval arrestb . | Sterile phenotype(% penetrance) . | Unc phenotype (severity) . | Vulval defectsc . |
|---|---|---|---|---|---|---|
| stu-8(oj1) | Moderate (mm) | + | 100 | Strong | P, V | |
| abc-1(oj2) | Nuclear division defective | Strong (mm) | +++ | 100 | Strong | P, V |
| spd-1(oj5) | Spindle defective/cytokinesis defective | Strong (mm) | + | >100 | Strong | P |
| zyg-1(oj7) | Centrosome duplication defective | Strong (mm) | ++ | 100 | Strong | V, P, M |
| stu-9(oj13) | Moderate (mm) | + | 100 | Weak | ||
| stu-10(oj14) | Abnormal spindle alignment | Moderate (mm) | + | 100 | Weak | P |
| stu-11(oj18) | Abnormal spindle alignment | Moderate (mm) | >100 | Weak | ||
| stu-12(oj21) | Moderate (mm) | + | >100 | Weak | P | |
| slo-1(oj23) | Slow development | Strong (mn) | + | >100 | Weak | |
| stu-13(oj24) | Strong (mn) | + | >100 | Weak | ||
| stu-14(oj26) | Moderate | ++ | >100 | Weak | P | |
| (mm) | ||||||
| stu-15(oj28) | Strong (mn) | + | 100 | Strong | P, M, V | |
| spd-2(oj29) | Abnormal microtubule organization | Strong (mm) | +++ | 100 | Strong | V, P |
| stu-16(oj30) | Moderate (mn) | + | >100 | Strong | P, V | |
| stu-17(oj31) | Moderate (mm) | >100 | Weak | V | ||
| stu-18(oj32) | Abnormal centrosome morphology | Moderate (mn) | +++ | >100 | Weak | M |
| stu-19(oj33) | Strong (mm) | + | 100 | Weak | ||
| cyk-2(oj34) | Cytokinesis defective | Moderate (mn) | + | 100 | Weak | |
| spd-3(oj35) | Abnormal microtubule organization | Strong (mm) | + | >100 | Weak | V |
| Mutation . | Cytological phenotype . | Emb phenotypea . | Larval arrestb . | Sterile phenotype(% penetrance) . | Unc phenotype (severity) . | Vulval defectsc . |
|---|---|---|---|---|---|---|
| stu-8(oj1) | Moderate (mm) | + | 100 | Strong | P, V | |
| abc-1(oj2) | Nuclear division defective | Strong (mm) | +++ | 100 | Strong | P, V |
| spd-1(oj5) | Spindle defective/cytokinesis defective | Strong (mm) | + | >100 | Strong | P |
| zyg-1(oj7) | Centrosome duplication defective | Strong (mm) | ++ | 100 | Strong | V, P, M |
| stu-9(oj13) | Moderate (mm) | + | 100 | Weak | ||
| stu-10(oj14) | Abnormal spindle alignment | Moderate (mm) | + | 100 | Weak | P |
| stu-11(oj18) | Abnormal spindle alignment | Moderate (mm) | >100 | Weak | ||
| stu-12(oj21) | Moderate (mm) | + | >100 | Weak | P | |
| slo-1(oj23) | Slow development | Strong (mn) | + | >100 | Weak | |
| stu-13(oj24) | Strong (mn) | + | >100 | Weak | ||
| stu-14(oj26) | Moderate | ++ | >100 | Weak | P | |
| (mm) | ||||||
| stu-15(oj28) | Strong (mn) | + | 100 | Strong | P, M, V | |
| spd-2(oj29) | Abnormal microtubule organization | Strong (mm) | +++ | 100 | Strong | V, P |
| stu-16(oj30) | Moderate (mn) | + | >100 | Strong | P, V | |
| stu-17(oj31) | Moderate (mm) | >100 | Weak | V | ||
| stu-18(oj32) | Abnormal centrosome morphology | Moderate (mn) | +++ | >100 | Weak | M |
| stu-19(oj33) | Strong (mm) | + | 100 | Weak | ||
| cyk-2(oj34) | Cytokinesis defective | Moderate (mn) | + | 100 | Weak | |
| spd-3(oj35) | Abnormal microtubule organization | Strong (mm) | + | >100 | Weak | V |
Letters in parentheses indicate genes in which maternal gene expression is required for viability (mm) and genes in which maternal or zygotic expression is sufficient for viability (mn).
Penetrance of larval arrest indicated as follows: +, 0-10%; + +, 10-20%; + + +, >20%.
Vulval defects are listed in order of highest penetrance. P, Pvl; V, Vul; M, Muv.
Vulval defects were evident among the nine lines with partially penetrant sterility. Again, the penetrance of vulval defects exhibited a positive correlation with the severity of the Stu phenotype. On the basis of appearance, the mutations spd-1 and stu-16 conferred the strongest Stu phenotypes; these mutations also had the highest penetrance of vulval defects (Table 2). All spd-1 hermaphrodites and many stu-16 hermaphrodites exhibited a Pvl phenotype. In contrast, the mutations stu-11, stu-12, slo-1, stu-13, stu-14, and stu-18 conferred mild Stu phenotypes and few vulval defects. Once again, a single exception to the correlation between these two phenotypes was noted: stu-17 hermaphrodites possessed a mild Stu phenotype and a high incidence of the Vul phenotype. These results suggest that, in general, the vulval lineages are affected to the same degree by these mutations as are the gonadal/germ-line lineages.
We also noted that two of the mutations conferred an additional phenotype. A higher than expected number of males were present among stu-10 and slo-1 adults. In a wild-type population, males (genetically XO) are produced spontaneously at an approximate frequency
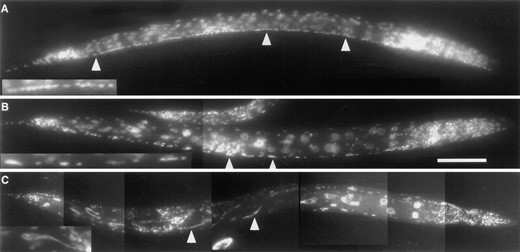
DAPI-stained wild-type and Stu mutant animals. Anterior is at right in all panels. (A) Wild type. Arrowheads indicate the single row of ventral cord nuclei that runs along the bottom edge of the animal. Inset, magnified view of nuclei. (B) zyg1(oj7). Arrowheads indicate polyploid nuclei within the ventral cord. Inset, magnified view of nuclei. (C) abc-1(oj2). Arrowheads indicate elongated intestinal cell nuclei. Inset, magnified view of an intestinal cell nucleus. Bar in B corresponds to 50 μm for A and 100 μm for B and C.
of one in 500 through nondisjunction of the X chromosome (Hodgkin et al. 1979). Among 56 stu-10 adults scored, 10 were male, and among 64 slo-1 adults, four were male. Unlike the other phenotypes scored in this test, this Him (high incidence of males) phenotype was almost certainly not because of a postembryonic defect but was instead because of defects in meiosis that occurred at the nonrestrictive temperature. Like the Him phenotype, the Stu phenotype may be because of chromosome missegregation; thus, stu-10 and slo-1 mutants may be defective in both meiotic and mitotic chromosome segregation.
To determine whether the Stu phenotype is a reliable indicator of postembryonic cell-division defects, we stained zyg-1 and abc-1 adults with the DNA-specific dye DAPI. Both zyg-1 and abc-1 animals contained many abnormal nuclei. In particular, we noted the presence of abnormal nuclei in the ventral nerve cord and intestine, two tissues in which postembryonic nuclear divisions occur (Sulston and Horvitz 1977). In wild-type animals, the adult complement of ventral cord nuclei is arranged in a longitudinal row that runs along the ventral midline. In DAPI-stained wild-type animals, these nuclei appeared of uniform size and intensity (Figure 2A). In contrast, in zyg-1 adults, many ventral cord nuclei were abnormally large and appeared to be polyploid, perhaps as a result of a nuclear-division defect. In abc-1 adults, intestinal cell nuclei exhibited an unusual morphology: nuclei were elongated and contained thin, wispy extensions (Figure 2C). These threads of DNA suggest a defect in chromosome segregation—and, as shown in Figure 4H, abc-1 embryos appeared to possess a related defect. In addition, zyg-1 and abc-1 animals possessed defects in gonadal and vulval nuclei, indicating that these mutations may lead to a general block in postembryonic cell division.
Stu mutations affect embryonic development: To determine to what extent these genes affect embryogenesis, we quantified the effect that each mutation had on embryonic viability. Homozygous mutant L4 larvae were shifted individually to 25°, and the percentage of viable embryos produced during two successive 24-hr periods was determined. For many strains, the percentage of viable embryos was higher during the first 24 hr than the second 24 hr, indicating that gene activity slowly decayed after the temperature shift. To avoid these confounding perdurance effects, we have chosen to consider the viability rate only during the second 24-hr period. These values are shown in Table 4.
Mutations ranged in their effects on embryonic viability. Nine mutations were found to strongly affect viability: hermaphrodites homozygous for abc-1, zyg-1, stu-15, spd-2, stu-19, or spd-3 did not produce any viable progeny, while spd-1, slo-1, and stu-13 mothers produced only a few survivors. Perhaps not surprisingly, this group contained many of the mutations with the strongest Stu phenotypes. Given the strong embryonic and postembryonic phenotypes of mutations abc-1, zyg-1, stu-15, and spd-2, it seems likely these mutations affect genes that are essential for development of many tissues.
Embryonic lethal phenotypes: Penetrance and parental tests
| Mutation . | P [m/m]aViable embryos (%) (n) . | R [m/m × +/+]bViable embryos (%) (n) . | S [m/+]cViable embryos (%) (n) . | |
|---|---|---|---|---|
| N2 | 97.3 (980) | NAd | NA | |
| stu-8(oj1) | 62.2 (24l) | 70.6 (357) | 93.1 | (404) |
| abc-1(oj2) | 0.0 (277) | 0.0 (352) | 96.3 | (734) |
| spd-1(oj5) | 0.4 (820) | 1.7 (462) | 97.1 | (513) |
| zyg-1(oj7) | 0.0 (63l) | 0.0 (606) | 89.6 | (632) |
| stu-9(oj13) | 36.6 (298) | 24.3 (325) | 97.2 | (797) |
| stu-10(oj14) | 55.8 (727) | 32.6 (436) | 96.8 | (888) |
| stu-11(oj18) | 45.8 (714) | 51.7 (354) | 96.8 | (524) |
| stu-12(oj21) | 37.5 (267) | 42.9 (268) | 94.8 | (688) |
| slo-1(oj23) | 0.7 (305) | 30.5 (430)e | 95.5 | (797) |
| stu-13(oj24) | 4.1 (339) | 44.6 (547)e | 94.2 | (466) |
| stu-14(oj26) | 75.5 (347) | 75.3 (364) | 97.9 | (334) |
| stu-15(oj28) | 0.0 (261) | 52.2 (288) | 97.7 | (963) |
| spd-2(oj29) | 0.0 (255) | 0.0 (264) | 94.7 | (732) |
| stu-16(oj30) | 31.8 (42l)f | 96.1 (337)e | 97.0 | (405) |
| stu-17(oj31) | 74.6 (315) | 86.3 (526) | 97.5 | (868) |
| stu-18(oj32) | 11.9 (404) | 53.1 (386)e | 97.8 | (558) |
| stu-19(oj33) | 0.0 (503) | 0.0 (258) | 97.9 | (707) |
| cyk-2(oj34) | 20.6 (315) | 39.3 (267)e | 99.1 | (352) |
| spd-3(oj35) | 0.0 (464) | 0.0 (342) | 99.3 | (738) |
| Mutation . | P [m/m]aViable embryos (%) (n) . | R [m/m × +/+]bViable embryos (%) (n) . | S [m/+]cViable embryos (%) (n) . | |
|---|---|---|---|---|
| N2 | 97.3 (980) | NAd | NA | |
| stu-8(oj1) | 62.2 (24l) | 70.6 (357) | 93.1 | (404) |
| abc-1(oj2) | 0.0 (277) | 0.0 (352) | 96.3 | (734) |
| spd-1(oj5) | 0.4 (820) | 1.7 (462) | 97.1 | (513) |
| zyg-1(oj7) | 0.0 (63l) | 0.0 (606) | 89.6 | (632) |
| stu-9(oj13) | 36.6 (298) | 24.3 (325) | 97.2 | (797) |
| stu-10(oj14) | 55.8 (727) | 32.6 (436) | 96.8 | (888) |
| stu-11(oj18) | 45.8 (714) | 51.7 (354) | 96.8 | (524) |
| stu-12(oj21) | 37.5 (267) | 42.9 (268) | 94.8 | (688) |
| slo-1(oj23) | 0.7 (305) | 30.5 (430)e | 95.5 | (797) |
| stu-13(oj24) | 4.1 (339) | 44.6 (547)e | 94.2 | (466) |
| stu-14(oj26) | 75.5 (347) | 75.3 (364) | 97.9 | (334) |
| stu-15(oj28) | 0.0 (261) | 52.2 (288) | 97.7 | (963) |
| spd-2(oj29) | 0.0 (255) | 0.0 (264) | 94.7 | (732) |
| stu-16(oj30) | 31.8 (42l)f | 96.1 (337)e | 97.0 | (405) |
| stu-17(oj31) | 74.6 (315) | 86.3 (526) | 97.5 | (868) |
| stu-18(oj32) | 11.9 (404) | 53.1 (386)e | 97.8 | (558) |
| stu-19(oj33) | 0.0 (503) | 0.0 (258) | 97.9 | (707) |
| cyk-2(oj34) | 20.6 (315) | 39.3 (267)e | 99.1 | (352) |
| spd-3(oj35) | 0.0 (464) | 0.0 (342) | 99.3 | (738) |
The percentage of viable embryos produced at 25° by homozygous mutant hermaphrodites. The number of embryos scored is indicated in parentheses. All animals were exposed to the restrictive temperature at the L4 larval stage except for stu-9 animals, which were exposed as young adults.
The percentage of viable embryos produced at 25° by homozygous mutant hermaphrodites mated to wild-type males. The number of embryos scored is indicated in parentheses. All hermaphrodites were exposed to the restrictive temperature at the L4 larval stage, except for stu-9 animals, which were exposed as young adults.
The percentage of viable embryos produced by heterozygous hermaphrodites at 25°. The number of embryos scored is indicated in parentheses. All animals were exposed to the restrictive temperature at the L4 larval stage.
NA, not applicable.
R-test value is significantly greater than corresponding P-test value (P > 0.05 by Student's t-test).
A significant number of the animals that escaped embryonic lethality arrested at the L1 stage. In these tests, only 0.5% of the stu-16 animals and 0.7% of stu-18 animals were viable beyond the L1 stage.
Embryonic lethal phenotypes: Penetrance and parental tests
| Mutation . | P [m/m]aViable embryos (%) (n) . | R [m/m × +/+]bViable embryos (%) (n) . | S [m/+]cViable embryos (%) (n) . | |
|---|---|---|---|---|
| N2 | 97.3 (980) | NAd | NA | |
| stu-8(oj1) | 62.2 (24l) | 70.6 (357) | 93.1 | (404) |
| abc-1(oj2) | 0.0 (277) | 0.0 (352) | 96.3 | (734) |
| spd-1(oj5) | 0.4 (820) | 1.7 (462) | 97.1 | (513) |
| zyg-1(oj7) | 0.0 (63l) | 0.0 (606) | 89.6 | (632) |
| stu-9(oj13) | 36.6 (298) | 24.3 (325) | 97.2 | (797) |
| stu-10(oj14) | 55.8 (727) | 32.6 (436) | 96.8 | (888) |
| stu-11(oj18) | 45.8 (714) | 51.7 (354) | 96.8 | (524) |
| stu-12(oj21) | 37.5 (267) | 42.9 (268) | 94.8 | (688) |
| slo-1(oj23) | 0.7 (305) | 30.5 (430)e | 95.5 | (797) |
| stu-13(oj24) | 4.1 (339) | 44.6 (547)e | 94.2 | (466) |
| stu-14(oj26) | 75.5 (347) | 75.3 (364) | 97.9 | (334) |
| stu-15(oj28) | 0.0 (261) | 52.2 (288) | 97.7 | (963) |
| spd-2(oj29) | 0.0 (255) | 0.0 (264) | 94.7 | (732) |
| stu-16(oj30) | 31.8 (42l)f | 96.1 (337)e | 97.0 | (405) |
| stu-17(oj31) | 74.6 (315) | 86.3 (526) | 97.5 | (868) |
| stu-18(oj32) | 11.9 (404) | 53.1 (386)e | 97.8 | (558) |
| stu-19(oj33) | 0.0 (503) | 0.0 (258) | 97.9 | (707) |
| cyk-2(oj34) | 20.6 (315) | 39.3 (267)e | 99.1 | (352) |
| spd-3(oj35) | 0.0 (464) | 0.0 (342) | 99.3 | (738) |
| Mutation . | P [m/m]aViable embryos (%) (n) . | R [m/m × +/+]bViable embryos (%) (n) . | S [m/+]cViable embryos (%) (n) . | |
|---|---|---|---|---|
| N2 | 97.3 (980) | NAd | NA | |
| stu-8(oj1) | 62.2 (24l) | 70.6 (357) | 93.1 | (404) |
| abc-1(oj2) | 0.0 (277) | 0.0 (352) | 96.3 | (734) |
| spd-1(oj5) | 0.4 (820) | 1.7 (462) | 97.1 | (513) |
| zyg-1(oj7) | 0.0 (63l) | 0.0 (606) | 89.6 | (632) |
| stu-9(oj13) | 36.6 (298) | 24.3 (325) | 97.2 | (797) |
| stu-10(oj14) | 55.8 (727) | 32.6 (436) | 96.8 | (888) |
| stu-11(oj18) | 45.8 (714) | 51.7 (354) | 96.8 | (524) |
| stu-12(oj21) | 37.5 (267) | 42.9 (268) | 94.8 | (688) |
| slo-1(oj23) | 0.7 (305) | 30.5 (430)e | 95.5 | (797) |
| stu-13(oj24) | 4.1 (339) | 44.6 (547)e | 94.2 | (466) |
| stu-14(oj26) | 75.5 (347) | 75.3 (364) | 97.9 | (334) |
| stu-15(oj28) | 0.0 (261) | 52.2 (288) | 97.7 | (963) |
| spd-2(oj29) | 0.0 (255) | 0.0 (264) | 94.7 | (732) |
| stu-16(oj30) | 31.8 (42l)f | 96.1 (337)e | 97.0 | (405) |
| stu-17(oj31) | 74.6 (315) | 86.3 (526) | 97.5 | (868) |
| stu-18(oj32) | 11.9 (404) | 53.1 (386)e | 97.8 | (558) |
| stu-19(oj33) | 0.0 (503) | 0.0 (258) | 97.9 | (707) |
| cyk-2(oj34) | 20.6 (315) | 39.3 (267)e | 99.1 | (352) |
| spd-3(oj35) | 0.0 (464) | 0.0 (342) | 99.3 | (738) |
The percentage of viable embryos produced at 25° by homozygous mutant hermaphrodites. The number of embryos scored is indicated in parentheses. All animals were exposed to the restrictive temperature at the L4 larval stage except for stu-9 animals, which were exposed as young adults.
The percentage of viable embryos produced at 25° by homozygous mutant hermaphrodites mated to wild-type males. The number of embryos scored is indicated in parentheses. All hermaphrodites were exposed to the restrictive temperature at the L4 larval stage, except for stu-9 animals, which were exposed as young adults.
The percentage of viable embryos produced by heterozygous hermaphrodites at 25°. The number of embryos scored is indicated in parentheses. All animals were exposed to the restrictive temperature at the L4 larval stage.
NA, not applicable.
R-test value is significantly greater than corresponding P-test value (P > 0.05 by Student's t-test).
A significant number of the animals that escaped embryonic lethality arrested at the L1 stage. In these tests, only 0.5% of the stu-16 animals and 0.7% of stu-18 animals were viable beyond the L1 stage.
Moderate effects on viability were observed for stu-8, stu-9, stu-10, stu-11, stu-12, stu-14, stu-16, stu-17, stu-18, and cyk-2; between 10 and 75% of the embryos produced by these strains hatched. In the cases of stu-16 and stu-18, the development of most of the survivors was arrested shortly after hatching; only 0.5% of stu-16 animals and 0.7% of stu-18 animals were viable beyond the first larval stage (Table 4). Many of the mutations with an incomplete Emb phenotype also had a mild Stu phenotype. These mutations might be hypomorphs, the weak phenotypes being because of residual gene activity. Alternatively, some mutations might be nulls: the corresponding genes might have partially redundant functions or play nonessential roles. Gene-dosage studies will be needed to address these possibilities. Nevertheless, as judged by their effects on early and late developmental events, these mutations probably define genes that function in many lineages.
Parental tests: All 19 Stu mutations conferred an Emb phenotype, suggesting that these genes might function throughout development. During the first several rounds of division, development of the embryo is largely controlled by maternal transcripts and proteins (Wood 1988). As a consequence, it is the genetic makeup of the mother, and not that of the embryo, that is relevant to this period of development. We performed two parental tests to determine to what extent these Emb phenotypes were influenced by maternal genotypes.
To determine whether the embryonic lethality associated with any of these 19 Stu mutations was a strict consequence of maternal genotype, we performed a male rescue (R) test (Wood et al. 1980). In the R test, a homozygous mutant hermaphrodite is mated with wild-type males at the restrictive temperature. The percentage of viable embryos produced by mated hermaphrodites is determined and compared to that of unmated controls. A significant increase in viability among the test embryos would indicate that maternal expression of the gene is not necessary; that is, the wild-type allele carried by the heterozygous offspring could suffice.
To determine whether maternal expression was sufficient for embryogenesis, we employed the S test (Wood et al. 1980). In the S test, viability is measured among the self-progeny of hermaphrodites heterozygous for the mutation of interest. All of the progeny will be supplied with wild-type maternal product. However, 1/4 of the progeny of a heterozygous mother will be homozygous, and thus unable to express wild-type gene product zygotically. Hence, the survival rate of this population indicates the degree to which maternally supplied gene products play a role in embryogenesis.
The results of the S test were striking in their uniformity: animals heterozygous for any one of the Stu mutations produced significantly more than 75% viable embryos (Table 4). Thus, in all cases, maternal gene activity was sufficient to allow most of the homozygous offspring to survive. While it was clear from the results of the S test that many zyg-1 homozygotes survived to hatching, mothers heterozygous for zyg-1 still produced moderate amounts (10.4%) of inviable offspring. When we examined the surviving progeny, we found that Stu animals still accounted for 25% of the offspring. As the homozygous offspring were not disproportionately affected, the lethality may be interpreted as a semidominant maternal effect.
In contrast to the results of the S test, those of the R test revealed differences among the Stu mutations. For 13 lines, stu-8, abc-1, spd-1, zyg-1, stu-9, stu-10, stu-11, stu-12, stu-14, spd-2, stu-17, stu-19, and spd-3, mating mutant hermaphrodites to wild-type males did not significantly increase embryonic viability (Table 4). For five of these genes, abc-1, zyg-1, spd-2, stu-19, and spd-3, the results were clear-cut: mated and unmated hermaphrodites did not produce any viable progeny. These genes exhibit a strict requirement for maternal expression, suggesting that they function early in development. In fact, as noted in the following section, four of these five mutants were found to exhibit defects during the first several rounds of cell division. For the other eight mutants, mated and unmated hermaphrodites produced similar nonzero levels of viable offspring, indicating that embryonic lethality is not rescued strongly—if at all.
For six mutations, maternal expression was found not to be necessary for embryonic viability. When mated to wild-type males, slo-1, stu-13, stu-15, stu-16, stu-18, and cyk-2 hermaphrodites produced significantly more viable progeny than unmated controls (Table 4). The lack of strict maternal effects suggests that these genes may not be necessary for the earliest period of development, before zygotic transcription is activated. Consistent with this hypothesis, we have not detected any serious defects during the first several cell cycles of stu-13, stu-15, stu-16, or stu-18 embryos. slo-1 and cyk-2 did confer early defects (see below), but in these cases the defects were somewhat variable, and the increases in viability observed in the rescue tests may be accounted for by the number of animals that escaped these early defects.
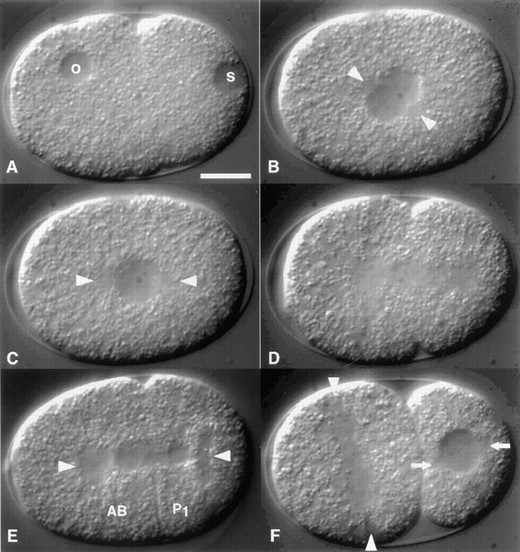
Stu mutant embryos possess cell-division defects: To determine what cytological defects were associated with these mutations, we analyzed the early divisions of mutant embryos. Mutant L4 larvae or adults were shifted to the restrictive temperature and allowed to produce embryos for approximately 16 to 24 hr. Young embryos were isolated from gravid mothers, and multiple-focal-plane time-lapse (i.e., 4D) data sets of the first several rounds of division were constructed. As the actual defects observed could sometimes vary between embryos, we recorded a minimum of three embryos from each mutant line. We report here only the defects observed multiple times. For purposes of comparison, early development of a wild-type embryo is shown in Figure 3.
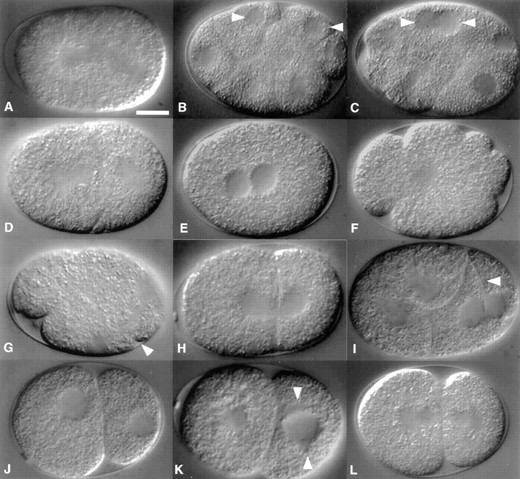
Ten of the mutants showed striking defects during the early cleavages (Figure 4), with the most common defect being cell-division failure. Embryos produced by mothers homozygous for spd-1, zyg-1, spd-2, cyk-2, or spd-3 exhibited reproducible failure of cell cleavage. These mutants could be divided into two groups on the basis of spindle morphology: spd-1, zyg-1, spd-2, and spd-3 embryos possessed abnormal mitotic spindles, while the spindles of cyk-2 embryos appeared normal. Among the other mutants, one exhibited a defect in nuclear division (abc-1), two exhibited defects in the positioning of the mitotic spindle (stu-10 and stu-11), one exhibited a defect in spindle morphology not associated with cytokinesis failure (stu-18), and another exhibited a defect in the timing of developmental events (slo-1). A more detailed description of the cytological phenotypes of these 10 mutants follows.
The spd-1, spd-2, spd-3, and zyg-1 mutations affect the mitotic spindle: Embryos produced by spd-1 mothers exhibited a defect in the late stages of cytokinesis. Some cells initiated furrowing normally but failed in the final pinching off of the membrane. Indeed, in those divisions that ultimately failed, cytokinesis seemed to be complete before regression of the furrow (Figure 4, B and C). These embryos also exhibited a defect in the behavior of the mitotic spindle. Through most of mitosis it appeared normal, but late in anaphase it broke at the midzone into two half-spindles (Figure 4A). While it is possible that the cytokinesis defect is a direct consequence of the spindle defect, we have not yet investigated the relationship between these two events.
Most zyg-1 embryos appeared normal through interphase of the second cell cycle. Invariably, however, both blastomeres of two-cell embryos exhibited an abortive mitosis and failed to divide (Figure 4D). Neither a metaphase plate nor a bipolar spindle was apparent following nuclear envelope breakdown. Instead, the DNA remained as a single large mass in the center of the cell throughout mitosis, ultimately being incorporated into an odd number of small nuclei. A smaller fraction of the embryos exhibited a similar pattern of defects during the first cell cycle. In all embryos, nuclear envelopes continued to disassemble and re-form at regular
Early embryonic development of C. elegans. (A) Pronuclear migration. The oocyte pronucleus (o) is traveling toward the sperm pronucleus (s) at the posterior of the embryo. A pseudocleavage furrow (visible at mid-egg length) forms but does not complete and regresses shortly after the oocyte pronucleus passes through it. (B and C) Rotation of the centrosome-pronucleus complex. The two pronuclei meet at the posterior then move to the center, where they undergo a 90° rotation to position the centrosomes (arrowheads) on the anterior-posterior axis. (D and E) First cleavage. The spindle initially forms at the center of the cell but becomes eccentrically placed toward the posterior during anaphase. The ensuing furrow divides the cell into a larger anterior cell (AB) and a smaller posterior cell (P1). Note that the AB centrosome is spherical, while the P1 centrosome is disc shaped (arrowheads in E). (F) Second division. The AB spindle forms first and is perpendicular to that of P1. The different spindle orientations are a consequence of different patterns of centrosome movements (Hyman and White 1987). The centrosome pairs of both AB and P1 are initially aligned along the same transverse axis—where, in AB, they establish the poles (arrowheads) of the mitotic spindle. However, prior to spindle formation in P1, the centrosomes (arrows), together with the nucleus, rotate by 90° so that the P1 spindle forms on the anterior-posterior axis. Anterior is to the left in all panels. Bar, 10 μm.
intervals, indicating that the cell cycle progressed unabated. Closer inspection of the time-lapse data sets revealed a defect in centrosome duplication or separation. In all cases, an aberrant mitosis was preceded in interphase by the formation of a single, abnormally large microtubule organizing center (MTOC). Typically, centrosomes are visible by means of DIC optics as granule-free regions associated with nuclei. This MTOC failed to resolve into two daughter centrosomes and remained positioned on the previous division axis. It was not clear from the DIC images whether the centrosome had divided, but under immunofluorescence microscopy, microtubules appeared to be organized around a single large centrosome (data not shown), consistent with a defect in duplication.
Failure to assemble a mitotic spindle was also observed in spd-2 embryos (Figure 4, E and F). As in zyg-1 embryos, the mass of DNA remained in the center of the cell, while the embryos continued to cycle through periods of nuclear envelope breakdown and re-formation (Figure 4F).
Representative cytological defects of Stu mutant embryos. (A) An spd-1(oj5) embryo with an abnormal anaphase spindle. In six of six spd-1 embryos observed, the first mitotic spindle broke at the midpoint into two half-spindles. (B and C) In five of eight spd-1 embryos, a late-stage cell-division defect was observed. A representative case is shown in B. Two daughter-cell nuclei (arrowheads) of an spd-1 embryo are separated by a nearly complete cytokinetic furrow. Nine minutes later (C) the furrow has regressed, and the daughter nuclei lie apposed. (D) Aberrant spindle assembly in a zyg-1(oj7) embryo. The nuclei in both cells have broken down, but monopolar rather than bipolar structures appear to have formed. Of eight zyg-1 embryos observed, three exhibited a similar defect during the first cell cycle and five during the second. (E) All spd-2(oj29) zygotes exhibit early developmental defects. The two pronuclei meet near the center of the embryo, and centrosomes are not apparent. (F) Failure to assemble a mitotic spindle in an spd-2 embryo during first mitosis. In all six spd-2 embryos observed, a mitotic spindle was absent. In the embryo shown, the DNA is visible as a clearing at the center of the cell, and multiple cytokinetic furrows can be seen at the periphery. (G) Abnormal first mitosis of a spd-3(oj35) embryo. The spindle is closely associated with the posterior cortex. A furrow is visible at the anterior end and is approximately parallel to the spindle. A second furrow can be seen to bisect the spindle (arrowhead). Similar defects were observed in all 11 spd-3 embryos examined. (H) Nuclear bridge of a two-cell abc-1(oj2) embryo. The re-forming nuclei lie close to the former cleavage plane and appear connected. Of five abc-1 embryos observed, all exhibited a similar defect. (I) Cytokinesis failure in a cyk-2(oj34) embryo. Two apposed daughter nuclei can be seen at the posterior of the embryo. An incomplete furrow (arrowhead) is visible nearby. Incomplete cytokinesis was observed in seven of 14 cyk-2 embryos. (J) Abnormal nuclear positioning as observed in seven of 12 cyk-2 embryos. (K) A stu-10(oj14) embryo in which the P1 centrosome-nucleus complex has failed to rotate. The AB spindle is not completely visible in this focal plane. The centrosomes are indicated by arrowheads. Abnormal alignment of the P1 centrosome-nucleus complex was observed in four of five stu-10 embryos and three of six stu-11 embryos. (L) Abnormal centrosome morphology in a two-cell stu-18(oj32) embryo. In 11 of 12 stu-18 embryos examined, the P1 centrosome did not flatten to the extent observed in wild-type embryos. Bar, 10 μm.
The defect in spindle morphogenesis was accompanied by several other defects, the most obvious of which was the apparent absence of an MTOC. In contrast to wild-type zygotes, in which the pronuclei-associated centrosomes are observable by means of DIC optics (Figure 3, B and C, arrowheads), such structures were not visible in spd-2 zygotes. The pronuclei of these embryos also exhibited defects in a number of microtubule-associated movements (Strome and Wood 1983; Hyman 1989; Hyman and White 1987). That is, the sperm pronucleus did not remain anchored at the posterior pole during pronuclear migration, but instead moved to the center of the zygote like the acentrosomal female pronucleus. Once apposed, the two pronuclei did not rotate by 90°, as occurs in wild-type embryos (compare Figures 3B and 4E). Thus, spd-2 embryos exhibited a number of defects, all of which can be attributed to a defect in microtubule organization.
Embryos produced by spd-3 mothers exhibited several defects. Polar bodies were often not properly extruded, leaving several maternally derived nuclei at the anterior of the zygote. During the ensuing interphase, these sister pronuclei co-migrated toward the sperm pronucleus at the posterior of the embryo. Mitotic spindles were difficult to detect by means of DIC optics, but in all embryos examined, the first spindle appeared to form at an angle to the anterior-posterior (A-P) axis and remained tightly associated with the posterior cortex throughout mitosis (Figure 4G). Two furrows were often observed: a unilateral furrow that bisected the spindle and a pseudocleavage furrow at some distance from and parallel to the spindle. Typically, first division failed, resulting in a multinucleate embryo.
The abc-1 mutation affects chromosome segregation: Embryos produced by abc-1 mothers possessed what appeared to be morphologically normal spindles. However, during telophase it became evident that the DNA had not segregated properly. The re-forming daughter nuclei remained connected to one another by a bridge that spanned the final connection between the daughter cells (Figure 4H). This connection, however, did not affect cleavage, as the furrow did not regress. Following cleavage, daughter nuclei often remained tightly associated with the former division plane. The connection between daughter nuclei is similar to that described for topoisomerase II mutants of yeast (Holm et al. 1985; Uemura et al. 1987) and may be explained by a failure to decatenate sister chromatids.
The cyk-2 mutation affects cytokinesis: About half of the embryos produced by cyk-2 mothers possessed a defect in the late stages of cytokinesis (Figure 4I). Spindle morphology appeared normal, and blastomeres underwent nuclear division. Cytokinetic furrows formed but progressed slowly and often failed to partition daughter nuclei. Incomplete furrows were seen to regress. Many cyk-2 embryos exhibited other defects as well. The positions of many nuclei were unstable and eccentric (Figure 4J), and the cytoplasm had an abnormal appearance, with granules exhibiting increased Brownian motion. cyk-2 blastomeres often lacked the well-defined contours of their wild-type counterparts, and the mutant embryos expanded to fill the entire volume of the egg-shell.
The stu-10 and stu-11 mutations affect spindle positioning: Two of the mutants exhibited abnormal positioning of the early mitotic spindles. The mutations stu-10 and stu-11 caused similar incompletely penetrant defects at the two-cell stage (Figure 4K). In 80% of stu-10 mutants and 50% of stu-11 mutants, the centrosome-nucleus complex of the posterior cell, P1, failed to rotate onto the A-P axis and remained in a transverse orientation until late in the cell cycle, when it appeared to be pushed onto the A-P axis by the elongating spindle of the anterior cell, AB. In both cases, the centrosome-nucleus complex was closely associated with the anterior P1 cortex. Although in both mutants this was the most common defect, it was not the only one: in a few stu-10 and stu-11 embryos, the centrosome-pronucleus complex did not complete rotation before first mitosis.
The stu-18 mutation affects centrosome morphology: The only visible defect in stu-18 embryos was a failure of the posterior spindle pole to undergo a shape change at the end of the first asymmetric division. In wild-type embryos, the centrosome inherited by the smaller P1 cell flattens from a sphere into a disc, whereas that of the larger AB cell remains spherical (Figure 3E). The significance of the shape change is not known, but it may be related to the close proximity of this centrosome to the cell cortex. In 11 out of 12 stu-18 embryos examined, the posterior centrosome failed to flatten completely (Figure 4L). We are not certain of the immediate consequences of such a defect and how the defect relates to the embryonic lethality of this strain, as stu-18 embryos continued to divide normally during the period of observation. Nonetheless, we find this phenotype intriguing, as centrosome flattening appears to be a common feature of unequal cell divisions (Dan 1979; Dan and Ito 1984).
The slo-1 mutation affects the rate of development: slo-1 embryos exhibited a phenotype completely different from that of the other mutants. These embryos appeared to develop normally but at a much slower rate than wild type. The longer the mother spent at 25°, the more slowly the embryos appeared to develop. Mothers that spent the longest periods at 25° produced embryos whose development was arrested during the first few cell cycles. While development seemed otherwise normal, we were unable to rescue one of these animals by returning it to low temperature.
Mutants not exhibiting early defects: Although examined extensively, stu-8, stu-9, stu-12–stu-17, and stu-19 embryos did not exhibit reproducible cytological defects during the early divisions. Many of these mutations had only moderate effects on embryonic viability, and thus the absence of a recurring defect was not surprising. On the other hand, several mutants had very strong (stu-13) or absolute (stu-15 and stu-19) Emb phenotypes yet failed to exhibit an early defect. It is possible that for these strains, the actual defects are not detectible by means of light microscopy. However, in the cases of stu-13 and stu-15, maternal gene activity was not found to be necessary, indicating that these genes are not required for early development. In the current study, we have analyzed mutant embryos only during the first several rounds of division. Analysis of the later stages of embryogenesis, including descriptions of the terminal phenotypes, might be helpful in determining the cytological basis for these cases of embryonic lethality.
DISCUSSION
Identification of new cell division genes: We have described the isolation and initial characterization of a set of ts cell-division mutants in C. elegans. Our approach was based in part on the phenotypes exhibited by two cell-division mutations described previously. The lin-5(e1348) and lin-6(e1466) mutations block virtually all postembryonic cell divisions—the principal exceptions being a few rounds of division in the germ line—including those of the gonad and ventral nerve cord (Albertson et al. 1978; Horvitz and Sulston 1980; Sulston and Horvitz 1981). While these two mutations affect different aspects of cell division, they confer similar Stu phenotypes, and neither affects viability. Thus, we reasoned that the Stu phenotype could be used to identify cell-division mutants while avoiding other types of lethal mutants. Another common property shared by the lin-5 and lin-6 mutations is that neither affects the embryonic divisions. While there are several possible explanations for this specificity, it seems likely that the lack of embryonic defects reflects maternal rescue of those cell divisions. As our data show, all of the mutants we isolated can be maternally rescued, indicating that most embryonic cell-division functions are maternally provided.
In formulating our approach, we decided that it would be most advantageous to screen for ts mutations. We wanted to identify cell-division genes by screening for mutants with a Stu phenotype, but we did not want to analyze postembryonic cell-division defects. Instead, we preferred to study the effects of cell-division mutations on the early embryonic divisions. Conditional alleles would allow us to inhibit maternally supplied gene activities and analyze the cytological effects of these mutations during this period. As the genes we were interested in were those encoding activities fundamental to proliferative and/or determinative cell division, we believed that the desired mutants would exhibit defects during all stages of the life cycle when cell division was occurring. Thus, we envisioned that the desired mutants would exhibit both an Emb and a Stu phenotype.
The results of the screen validate our approach. Nineteen ts mutations that conferred both Stu and Emb phenotypes were identified (Table 3). When analyzed for cytological defects, many mutants were found to exhibit defects in cell division. These included failure to properly segregate DNA (abc-1), defects in placement (stu-10 and stu-11) and morphogenesis (zyg-1, spd-1, spd-2, and spd-3) of the mitotic spindle, defects in cytokinesis (spd-1, spd-2, spd-3, and cyk-2) and prolonged cell cycle times (slo-1). Although not all mutants were found to exhibit cell-division defects, all remained viable when shifted to the restrictive temperature postembryonically, arguing that none of the mutations affected essential metabolic functions. These results strongly support our assumption that the Stu phenotype can serve as a reliable indicator of cell-division defects. However, we do not believe the Stu phenotype to be absolutely specific to cell-division failure. Mutations affecting any developmental process required for formation of the gonad and ventral nerve cord could conceivably yield a Stu phenotype. For instance, genes with general roles in cell differentiation could be mutated to a Stu phenotype. Despite exhaustive analysis of a number of mutant lines (stu-8, stu-9, stu-12–stu-17, and stu-19), we were unable to detect early cell-division defects. It therefore seems probable that at least some of the Stu mutations affect developmental processes other than cell division; only further analysis of the embryonic and postembryonic developmental defects associated with these mutations will allow us to determine the types and frequencies of defects that give rise to this phenotype. Nonetheless, when combined with cytological analysis, the screen provides a very powerful approach to identifying cell-division mutations in C. elegans.
A measure of success of any screen is whether it has identified novel genes. A large number of conditional emb mutations already exist, and many of these confer gonadogenesis defects when exposed to the restrictive temperature postembryonically (Hodgkin 1997). While earlier screens were able to identify mutations that conferred both Emb and Stu (or sterile) phenotypes, they did not specifically focus on this group of polyphasic mutants (Hirsh and Vanderslice 1976; Miwa et al. 1980; Cassada et al. 1981). We therefore reasoned that a screen designed specifically to isolate such mutants would likely identify many new genes. Our results strongly support this notion: mapping and complementation tests have indicated that 17 of the 18 Stu mutations tested define novel genes.
Can we estimate the number of genes that can be mutated to a conditional Stu phenotype? As no two of the new mutations are allelic, we are unable to do so. However, the fact that we have not obtained multiple alleles of any of the 18 mapped genes suggests that the screen is far from saturation. In fact, none of the new mutations are allelic to lin-5 or lin-6 mutations or to any one of three ts Stu mutations identified in an earlier screen (Livingstone 1991).
Genetic analysis: Our results indicate that the new mutants are diverse with respect to their requirements for maternal expression. Embryonic lethality associated with 13 mutants behaved like a strict maternal effect. Five such mutants exhibited a complete Emb phenotype, and four of these possessed cell-division defects during the early cleavages—indicating, as expected, that genes with a strict maternal effect are likely to be required for the earliest cell divisions. In contrast, six Stu mutations were found to have a partial parental effect, either parental or zygotic expression being sufficient for embryogenesis. Of these, three were found to possess early defects (slo-1, stu-18, and cyk-2). In situ hybridization studies have indicated that at least some zygotic genes are transcribed as early as the four-cell stage (Seydoux and Fire 1994); these three partial parental-effect mutations with early defects may thus define genes that are expressed very early in development. On the other hand, for some of these genes, the absence of a strict maternal effect may be specific to the alleles tested; stronger alleles may behave like strict maternal-effect lethal mutations.
Remarkably, for all Stu mutations, tests to determine whether a wild-type maternal allele was sufficient for embryogenesis yielded nearly identical results. For each of the 19 mutants tested, embryonic lethality was strongly rescued by the maternally supplied gene product. This result was obtained irrespective of the strength of the mutation or of the process affected. Roughly 550 divisions occur during embryogenesis, and the results of the S test strongly suggest that all of these divisions are executed using predominantly maternally supplied factors. Our results have an important implication for identification of cell-division mutants in C. elegans; that is, many such mutants would likely not present as zygotic-effect embryonic lethals.
Cytological analysis: In analyzing the early divisions of mutant embryos, we found that many of the mutations recovered in this screen conferred intriguing cell-division defects. Although the mutation set was rather small, we did not notice a bias in the type of cell-division mutant isolated. The phenotypes observed were varied and included defects in chromosome segregation, spindle morphogenesis and alignment, centrosome duplication, and cytokinesis. Thus, as we hypothesized, our approach is capable of identifying genes with a broad range of cell-division functions.
The abc-1 mutation affects chromosome segregation: The abc-1 mutation was unique in that it was the only member of this set of mutations that appeared to primarily affect chromosome segregation. During anaphase in abc-1 embryos, a thin bridge connected the separating DNA complements. This “anaphase bridging” is similar to the phenotypes described for topoisomerase II mutants of yeast (Holm et al. 1985; Uemura et al. 1987) and barren (barr) mutations of Drosophila (Bhat et al. 1996). barr encodes a novel, chromosome-associated protein that interacts with Topoisomerase II and modulates its activity. During mitosis in barr embryos, centromeres move apart at anaphase, but sister chromatids remain linked, suggesting that Topisomerase II and Barren protein act to decatenate chromatids at anaphase. The phenotype of abc-1 mutants suggests that the product encoded by this gene may play a role in this process in C. elegans. We searched the GenBank Sequence Database for C. elegans Barren and Topoisomerase II homologues. While a Barren homologue was not detected, several sequences with similarity to Topoisomerase II were found, but none of these mapped in the vicinity of abc-1.
Four mutations affect spindle morphogenesis: A distinct and dramatic anaphase defect was observed in spd-1 mutant embryos. Although the spindle formed and behaved normally through most of mitosis, late in anaphase it bent at the midzone. Often it appeared to break, completely separating into two half-spindles. Associated with this phenotype was a late cytokinesis defect: furrows initiated normally but failed to complete. As not all cells that exhibited the spindle defect exhibited the cytokinesis defect, we are not sure of the relationship between these two defects. One possibility is that the cytokinesis defect is a direct consequence of disruption of the spindle midzone. Experiments in tissue-culture cells suggest that in at least some cell types, the midzone appears to be required for cytokinesis (Wheatley and Wang 1996). Similarly, mutations in the gene encoding the Drosophila kinesin-like protein KLP3A disrupt the central spindle in spermatocytes and lead to defects in cytokinesis (Williams et al. 1995).
The zyg-1 and spd-2 mutations blocked formation of a bipolar mitotic spindle. In zyg-1 embryos, the centrosomes failed to divide during the first or second round of division. In all cases, a single large MTOC was apparent. Under indirect immunofluorescence, this structure appeared as a single large focus of microtubules (data not shown) and not as two closely apposed foci. These observations are consistent with a defect in centrosome duplication rather than a defect in separation of daughter centrosomes.
Including oj7, there are six mutant alleles of the zyg-1 gene. Only the oj7 and b1 alleles cause Stu phenotypes, and in the case of b1, this phenotype is incompletely penetrant. All, however, cause defects in formation of the vulva and strong maternal-effect lethal phenotypes. Cytologically, all confer defects in centrosome duplication (C. Caron and K. Kemphues, personal communication). While all zyg-1 mutations affect duplication at the two-cell stage, it appears that only oj7 affects duplication in the zygote. Given the more severe embryonic and postembryonic phenotypes associated with oj7, it is likely to be the strongest member of this allelic series. Interestingly, two of the mutants often have multiple centrosomes associated with the sperm pronucleus, but these fail to duplicate after the first cell cycle (C. Caron and K. Kemphues, personal communication). Thus, the zyg-1 gene product appears to play a complex role in regulating the number of centrosomes.
The spd-2 mutation appeared to affect microtubule organization. In addition to the defect in spindle morphogenesis, two microtubule-dependent movements of the pronuclei were affected. In wild-type embryos, migration of the two pronuclei toward one another can be blocked with drugs that disrupt microtubules (Strome and Wood 1983). Although the spd-2 mutation did not block migration, it did disrupt the normal migration pattern; in wild-type embryos, the pronuclei travel unequal distances and meet near the posterior pole, while in spd-2 embryos the nuclei travel equal distances and meet in the center of the embryo. In wild-type embryos, the pronuclei also undergo a second microtubule-dependent movement. Prior to spindle morphogenesis, the two pronuclei and associated centrosomes rotate by 90° to position the first mitotic spindle on the A-P axis. In spd-2 embryos, the pronuclei do not rotate. These observations suggest that some aspect of the microtubule cytoskeleton is affected by the spd-2 mutation. Consistent with this idea, we have found by immunofluorescence microscopy that microtubules are poorly organized in spd-2 embryos (data not shown).
spd-3 embryos exhibited defects in polar-body formation, spindle positioning, and cytokinesis. The first spindle was typically transverse and misplaced toward the posterior of the zygote, where it often elicited a furrow midway between the poles. In addition, an ectopic furrow, parallel to the spindle, formed in the anterior. Often these furrows failed to complete. This phenotype bears some resemblance to that of zyg-9 mutants and to that of wild-type embryos that have been treated with the microtubule inhibitor nocodazole (Strome and Wood 1983; Kemphues et al. 1986). In these embryos, the first mitotic spindle is located at the posterior and oriented transversely, as in spd-3 embryos. The spindle of zyg-9 embryos—like that of nocodazole-treated embryos—is smaller than usual, indicating that the zyg-9 gene product affects microtubule stability. In contrast, the spindle of spd-3 embryos does not appear abnormally small, indicating perhaps that the spd-3 mutation affects microtubules in a manner different from that of zyg-9 and nocodazole.
Our observations of these new spindle-defective mutants indicate that the spindle plays at least two distinct roles in cytokinesis. While the spindle seems dispensable for the act of furrowing, it appears to be required to organize cortical contractile activity. The zyg-1 and spd-2 mutants lacked bipolar mitotic spindles but possessed contractile activity during mitosis. However, the cortical contractions were disorganized. In spd-2 embryos, furrowing was not restricted to a single cortical region (Figure 4F), and the multiple furrows that formed failed to complete. Thus, the mitotic apparatus seems to spatially regulate the cortical contractions, resulting in the formation of a single cytokinetic furrow that bisects the spindle. The spindle also seems to be required for the completion of cytokinesis. In spd-1 embryos, apparently normal bipolar spindles were observed, and each elicited a single furrow between the poles. However, many spindles tended to break at the midzone, and cytokinetic furrows—which appeared to completely separate daughter cells—often regressed. These observations suggest that the spindle midzone is required for the terminal phase of cytokinesis—possibly for the final pinching off of the membrane. Consistent with this idea, two furrows are often observed in nocodazole-treated embryos: one that bisects the spindle midzone and completes; and a second, not associated with the spindle, that does not complete (Hird and White 1993). Similar observations have been made during polar-lobe formation in molluscan eggs (Conrad et al. 1973), indicating that the spindle may be required for the completion of cytokinesis in many cell types.
The cyk-2 mutation affects cytokinesis: A late cytokinesis defect was observed in cyk-2 embryos. Furrows formed but ingressed slowly, such that at telophase, cell borders were incomplete. The effect of this mutation may be kinetic; that is, the cyk-2 may simply slow the progression of cytokinesis. This could lead to the presence of incomplete furrows when the cells reenter interphase. The interphase cytoplasm may not be suitable for cytokinesis, leading to relaxation of the furrow. A similar defect has been described for the keule mutant of Arabidopsis (Assaad et al. 1996).
Three mutations affect positioning or morphology of the centrosome: Several mutants exhibited defects not related to cytokinesis. The stu-10 and stu-11 mutations affected spindle positioning, most often at the two-cell stage. The phenotype was similar in both mutants: the P1 spindle formed on a transverse axis rather than on the A-P axis. Only very late in the cell cycle did the spindle take on the correct orientation, perhaps utilizing a backup mechanism. Several lines of evidence indicate that spindle positioning relies upon the interactions of astral microtubules with the cell cortex (Hyman and White 1987; Hyman 1989; Waddle et al. 1994; Cheng et al. 1995; Etemad-Moghadam et al. 1995). The stu-10 and stu-11 mutations may interfere with these interactions, leading to defects in alignment.
Although quite different, the phenotype exhibited by stu-18 embryos may be related to that of stu-10 and stu-11 embryos. In stu-18 embryos, the shape of the P1 centrosome does not change from that of a sphere to that of a flattened disc, as invariably happens in wild type. Neither the mechanism whereby this change occurs nor its significance is known. However, the change in centrosome morphology might, like spindle alignment, involve astral microtubule/cell cortex interactions, as some of the same mutations that affect spindle alignment affect centrosome morphology (Cheng et al. 1995). Thus, the stu-10, stu-11, and stu-18 mutations may affect different sets of these interactions.
The isolation of mutations affecting a specific biological function provides a very effective means of studying that function. Many of the mutants isolated in this study exhibited intriguing defects in cell division. Prominent among this group are those with defects in assembly and behavior of the mitotic spindle. We are interested in the roles played by these genes in spindle morphogenesis and believe that further genetic and cytological analysis of these spindle-defective mutants will provide us with a deeper understanding of this process.
Acknowledgement
We thank Lisa Kadyk and Kara Maxwell for comments on the manuscript, Judith Kimble and Ann Rose for generously providing strains, and Charles Thomas for providing support for 4D imaging. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources, National Institutes of Health (NIH). This work was supported by U.S. Public Health Service grant GM-52454. K.F.O. was supported by postdoctoral fellowship GM-17002 from the NIH.
Footnotes
Communicating editor: R. K. Herman
LITERATURE CITED