-
PDF
- Split View
-
Views
-
Cite
Cite
Patrick C Phillips, The Language of Gene Interaction, Genetics, Volume 149, Issue 3, 1 July 1998, Pages 1167–1171, https://doi.org/10.1093/genetics/149.3.1167
Close - Share Icon Share
The study of variation and heredity, in our ignorance of the causation of those phenomena, must be built of statistical data, as Mendel knew long ago; but, as he also perceived, the ground must be prepared by specific experiment. The phenomena of heredity and variation are specific questions. That is where our exact science will begin. Otherwise we may one day see those huge foundations of “biometry” in ruins.
W. Bateson (1902)
VERY soon after the rediscovery of Mendel it was realized that the multilocus nature of inheritance could not be understood solely by examining the action of individual genes and then predicting how these genes would behave in concert simply by combining the separate observations. Frequently genes interact with one another, distorting simple Mendelian ratios and sometimes leading to novel phenotypes. Ninety years ago, William Bateson coined the term “epistasis” to describe this sort of interaction (he appears to have first used the term in 1908, although it is most clearly spelled out in Bateson 1909). Epistasis translates directly to “standing upon,” and was meant to describe the situation in which the action of one locus masks the allelic effects at another locus, much in the same way that complete dominance (the way in which most genes appeared to work at the time) involves the masking of one allele by another. The locus being masked is said to be “hypostatic” to the other locus.
At the same time that he was investigating all manner of Mendelian segregation ratios, Bateson was involved in a heated battle with the Biometrical school of genetics, exemplified by the views of Karl Pearson and W. F. R. Weldon (Provine 1971). The Biometricians focused on the continuous range of variation among relatives in contrast to the discrete differences studied by the Mendelians. Eighty years ago, in one of the seminal papers in genetics, R. A. Fisher (1918) conclusively ended the debate by showing analytically that Mendelian segregation was compatible with the Biometrician's laws of heredity (something that had been argued qualitatively by a number of others before Fisher). In this same work, he also established several branches of statistics and laid the groundwork for the entire field now known as quantitative genetics. It was quite a paper.
The essential problem with which Fisher had to deal was how to map the discrete segregation of alleles onto the continuous range of measured traits. Fisher noted that in conducting this mapping, it was entirely possible (although, as it was later clear, not likely to his mind), that one would not be able to predict the quantitative phenotype of a particular two-locus genotype by simply adding the effects of two loci together. With undoubted allusion to Bateson's unusual segregation ratios, Fisher called these nonadditive interactions “epistacy.” This term rapidly became simply “epistasis,” and two related, but distinct meanings of the same word entered the geneticist's vocabulary (Wade 1992a).
It is perhaps telling that this duality in meaning rarely causes much confusion for those using the term. Mendelian and molecular geneticists tend to use epistasis in the strict sense of Bateson, while evolutionary and quantitative geneticists use epistasis to mean just about any form of gene interaction. After briefly tracing the history of the use of “epistasis,” I suggest that there is good reason to start thinking about eliminating this potential source of confusion. Some of the difficulty in choosing the right word to describe gene interactions results from the lingering conflict between the Biometrical and Mendelian views of characterizing gene action. The resolution to this difficulty points the way to the possible final synthesis of these two approaches.
EPISTASIS FROM THE MENDELIAN VIEWPOINT
epistasis 1. Genetics. An interaction between nonallelic genes, especially an interaction in which one gene suppresses the expression of another.
American Heritage Dictionary of the English Language (1992)
Although officially given a name some ninety years ago, the phenomenon of gene interaction was discovered a few years earlier. In their study of the genetics of chicken combs, Bateson and Punnett (in Bateson et al. 1905) noted that one type of comb, the single, was rarely produced in their crosses and that its presence was difficult to reconcile with a simple Mendelian genetic system. Making good use of Punnett's recently derived method of creating a matrix, a “Punnett square,” to describe all possible combinations of gametes, they concluded that comb inheritance could be described by Mendelian segregation of two factors, with the single phenotype appearing only in the rare double-recessive homozygotes. Punnett later found similar results in his parallel work on sweet peas (reviewed in Punnett 1920). Weinberg (1910) also noted the possibility of such interactions, calling them komplizierter polyhybridismus (complicated polyhybridisms). It is perhaps not surprising that Weinberg's term did not catch on, and his work unfortunately appears to have been largely ignored.
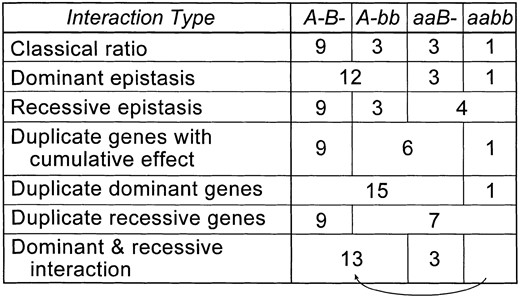
Despite Bateson's coining of a term to describe this situation and a great deal of coverage of unusual segregation ratios (usually described under the heading “interaction of factors”) in early textbooks on genetics, there appears to have been little use of the word “epistasis” in the first few decades of this century [even by Punnett (1920) himself; see also Castle (1926)]. By the thirties, however, the strength of the Mendelian approach was firmly established and its power illustrated through a description of all manner of segregation ratios, with epistasis being a prime example (e.g., Lindsey 1932; Sinnott and Dunn 1932; Snyder 1935). A menagerie of special cases was collected and given names (Figure 1).
Yet by the fifties, interest in epistasis from a Mendelian point of view appears to have waned substantially. Gene interaction and unusual segregation ratios received diminished attention in textbooks (e.g., Colin 1956), although newer editions of older texts still maintained wider coverage (Sinnott et al. 1958). Perhaps the decrease in interest in epistasis was because testing the Mendelian paradigm in more complex situations was no longer interesting. It was now self-evident that Mendelian segregation of chromosomes composed primarily of DNA provided an elegant physical mechanism for this segregation. Perhaps it was because variable mortality made Drosophila, still the dominant experimental system, not always the best organism for precise testing of complex segregation ratios. Most likely, geneticists at the time were busy getting on to other things.
Indeed, the next few decades were consumed by the molecular revolution. Since the eighties, however, the analysis of epistasis has made a strong comeback as an important means of ordering genes in developmental pathways (e.g., Avery and Wasserman 1992). This can sometimes include complex bifurcating pathways involving dozens of genes (e.g., Thomas 1993). The shift back toward epistasis is reflected in an expansion of coverage
in recent editions of introductory genetics textbooks (e.g., Weaver and Hedrick 1997).
It is clear that the term epistasis in the exact sense in which it was used by Bateson now plays a central role in modern genetics beyond the mere description of segregation ratios and well into the analysis of gene function. In the coming decade, gene interaction, in the very physical sense of the direct interaction between gene products that form molecular machines and signaling pathways, is likely to become more and more central to genetic analysis, and “epistasis” is equally likely to frequent the geneticist's lexicon.
EPISTASIS FROM THE STATISTICAL GENETICS VIEWPOINT
When we move from Mendelian to biometrical analysis we must retain the concepts which have proved successful and necessary at the old level; but we must be prepared to see them appear, and to use them, in the new way necessitated by the new level of integration at which we are working.
K. Mather (1949, p. 400)
The impact of Fisher's (1918) paper on the entire field of quantitative genetics has been immense. Almost all of the notation and methods of analysis used today can be traced directly to this point source. It is curious, then, that Fisher's derivative term, epistacy, has not been used to describe quantitative gene interaction. Sewall Wright, the geneticist most closely identified with championing the importance of gene interactions in evolutionary change, used neither term to describe gene interactions in his most famous papers (Wright 1931, 1932), although a few years later his use of “epistasis” was firmly established (Wright 1935).
The use of epistasis to describe gene interaction in quantitative genetics is ostensibly similar to Bateson's use in describing segregation ratios. The issue here is whether or not the phenotype of a given genotype can be predicted by simply adding (or multiplying, depending on scale) its component single-locus effects. To the extent that this cannot be done, then the leftover bits are called the epistatic deviations. This is much more inclusive than Bateson's original use because many forms of gene interaction can lead to epistatic deviations.
The overall complexity of the situation was sorted out by Cockerham (1954, 1956) and Kempthorne (1954). They broke the epistatic deviations into particular components that could be described by a statistical model, leading to terms like additive-by-additive, additive-by-dominance, and dominance-by-dominance epistasis (Cockerham 1954). Kempthorne (1954), from the British school, still used “epistacy” in this context, although in his influential book a few years later (Kempthorne 1957) he acknowledged that “epistasis” was the more commonly used descriptor. With this last gasp, the term “epistacy” appears to have winked out of existence.
More recent approaches to dealing with epistasis have focused on Fisher's (1918) most lasting contribution to statistical genetics, the idea that the effect of a gene needs to be estimated within the context of the population from which it is drawn. An alternative approach would be to have a specific model of how genes function and interact and then to construct a building-block description of the phenotype based on the genetic composition of the individual (Crow and Kimura 1970, pp. 77ff; Tachida and Cockerham 1989). A similar approach has been suggested by Cheverud and Routman (1995), in what they call physiological epistasis (in contrast to statistical epistasis). The problem with building-block models is that they are still statistical abstractions (as is made clear by Tachida and Cockerham 1989) and are thus really no closer to the actual physiological mechanism of gene interaction than the effects models. Building-block models are limited by the particular genotypes that happen to be under study (and the particular loci being examined within those genotypes), whereas Fisher was concerned that the total number of possible genotypes could rapidly outpace the total size of any population (see Fisher 1941). The advantage of Fisher's approach is that it yields the parameters that are important for describing evolutionary change while at the same time being relatively insensitive to the particular sample of genotypes under study. The disadvantage of this approach is that the molecular and physiological nature of the gene interactions themselves can be masked by statistical abstraction and vagaries of allele frequencies. Other formulations have been suggested (e.g., Schnell 1984; Wagner et al. 1998), but the best approach is undoubtedly going to be determined by the questions to be answered.
AN EXPANDED LANGUAGE OF GENE INTERACTIONS
epistasis 2. Medicine. A film that forms over the surface of a urine specimen. 3. The suppression of a bodily discharge or secretion.
American Heritage Dictionary of the English Language (1992)
Part of the difficulty in reconciling the Mendelian and statistical formulations of epistasis is that our language for describing gene interactions is currently far too limited. Not all gene interactions are equal from the standpoint of understanding gene action, particularly in their consequences for evolution at those loci (Whitlock et al. 1995; Fenster et al. 1997). In fact a number of different terms that describe gene interactions do not fit the expectations of classical epistasis and might be overlooked simply because they are not called by the expected name(see Figure 1). For example, compensatory and suppressor mutations clearly involve gene interactions (Phillips 1996; Stephan 1996) and may in fact describe the same type of interaction (it is rare for all possible genotypes to be investigated in these cases). Similarly, so-called synthetic phenotypes are sometimes observed when two mutations with apparently separate effects are combined in the same individual. The most extreme case of this is synthetic lethality (Dobzhansky 1946), which could potentially be of evolutionary importance (Phillips and Johnson 1998), but may be too rare to be significant (Temin et al. 1969; Thompson 1986). Among evolutionary biologists, differentiating reinforcing (or synergistic) from diminishing (or antagonistic) epistasis (Crow and Kimura 1970, pp. 80–81) is currently a hot topic because of their role in the evolution of sex and recombination (Kondrashov 1995). It is important to note that these forms of gene interaction are usually measured as average attributes (Otto 1997; Otto and Feldman 1997), which may be more illuminating after a careful analysis of the effects themselves (e.g., Clark and Wang 1997; Elena and Lenski 1997). Finally, the genetic incompatibility that drives speciation must be caused by gene interactions, the form of which can be quite complex (Wu and Palopoli 1994).
When discussing allelic interactions within a locus, it is clear that the consequences of complete dominance, partial dominance, underdominance, and overdominance are quite different, and we have particular terms to describe each situation. To use a single term, epistasis, to describe all gene interactions suggests that we either do not care or do not know how to deal with the complexity that interlocus interactions bring. Different forms of gene interaction have different consequences, and we should move toward elucidating the nature of these consequences and which forms of interaction are most prevalent.
For most of the 80 years since Fisher (1918), these sorts of considerations really did not matter, and the Mendelian and statistical definitions of epistasis could coexist because they rarely crossed paths. This period of separation is appearing to be near an end. Studies of quantitative trait loci (QTL) are beginning to bridge the gap between continuous variation within populations and the genetic mechanisms that generate that variation. In the few studies of this nature that have been conducted, genic interactions have been common (e.g., Doebly et al. 1995; Long et al. 1995; Routman and Cheverud 1997). With luck in a few particular circumstances, we will no longer be talking about statistical abstractions but instead will be dealing with real loci with particular effects. Especially interesting are studies that actually attempt to physically generate the genotypic decomposition of interactions by targeting particular loci and limiting total genotypic variation to just a few sites (Clark and Wang 1997; Elena and Lenski 1997). While it is possible that epistatic effects discovered using a mutational approach might be expected to be more common and perhaps more extreme as the genetic system is perturbed far from its normal state (Crow 1990), it appears likely that as more and more QTL are investigated, genic interactions will begin to come to the fore in statistical genetics much as they have as a tool for functional analysis in Mendelian genetics. Once freed from being relegated to “epistatic variance” as a nuisance term (with all of the statistical ambiguities that that entails; Wade 1992b), context and interaction may indeed become of the essence (Lewontin 1974). However, as Fisher long argued, even if the building blocks display gene interaction, the evolutionary implications of these interactions need not necessarily be profound. The resolution of this conflict awaits more data on the precise nature of the interactions themselves. The debate on the significance of gene interactions has needed the illumination of this sort of data for the last 80 years. It is exciting to note that we appear poised to collect it.
CONCLUSIONS
This definition of ‘epistatic’ (apparently first used by Fisher in 1918) is a bit more inclusive than what Bateson originally meant. Advancing knowledge ofthe multiplicity of possible kinds of non-additive interactions of genes has destroyed the distinctness of the original definition. If this broadening of the original definition proves too confusing, perhaps these epistatic deviations will ultimately be called merely ‘interactions’.
J. L. Lush (1940, p. 295)
As statistical and Mendelian genetics begin to slowly converge experimentally (as Fisher allowed them to do conceptually long ago) the question of what to call gene interactions becomes more than mere semantics. It is conceivable that sometime in the near future two articles published in Genetics will discuss possible epistasis between the same two loci and mean very different things by this, perhaps even coming to different conclusions. It would be a shame to allow semantics to be a barrier to what may well be the final unification of the Mendelian and Biometrical schools.
It is clear that Bateson's more restrictive use of “epistasis” has historical precedence and is still being used in this way by the majority of geneticists. (It is also surprisingly similar in form, if not nuance, to the medical definitions given above.) The onus is therefore on the statistical geneticists to refine their use of the word. One course of action would be to simply continue using the same word for both meanings, being very careful to allow context to determine which meaning is implied. Alternatively, statistical genetics could return to Fisher's (1918) original descriptor, epistacy. This seems unlikely and is only slightly less confusing. A final solution would be to heed Lush's advice and simply call the more general case what it is, gene interaction. This might make it clearer that simply detecting an interaction and giving it a general name is insufficient. The actual nature of the interaction is central and is the thing upon which arguments regarding the general significance of gene interactions can be built.
Acknowledgement
I thank Mike Whitlock for pointing out the dictionary definitions of epistasis. I also thank Mike Wade and Mike Whitlock for many helpful discussions over the years and Norman Johnson and Mike Whitlock for comments on the manuscript. Supported in part by National Science Foundation grant number DBI-9722921 and National Institutes of Health grant number GM54185.
LITERATURE CITED