-
PDF
- Split View
-
Views
-
Cite
Cite
Min Xu, Hargeet K Brar, Sehiza Grosic, Reid G Palmer, Madan K Bhattacharyya, Excision of an Active CACTA-Like Transposable Element From DFR2 Causes Variegated Flowers in Soybean [Glycine max (L.) Merr.], Genetics, Volume 184, Issue 1, 1 January 2010, Pages 53–63, https://doi.org/10.1534/genetics.109.107904
Close - Share Icon Share
Abstract
Active endogenous transposable elements, useful tools for gene isolation, have not been reported from any legume species. An active transposable element was suggested to reside in the W4 locus that governs flower color in soybean. Through biochemical and molecular analyses of several revertants of the w4-m allele, we have shown that the W4 locus encodes dihydroflavonol-4-reductase 2 (DFR2). w4-m has arisen through insertion of Tgm9, a 20,548-bp CACTA-like transposable element, into the second intron of DFR2. Tgm9 showed high nucleic acid sequence identity to Tgmt*. Its 5′ and 3′ terminal inverted repeats start with conserved CACTA sequence. The 3′ subterminal region is highly repetitive. Tgm9 carries TNP1- and TNP2-like transposase genes that are expressed in the mutable line, T322 (w4-m). The element excises at a high frequency from both somatic and germinal tissues. Following excision, reinsertions of Tgm9 into the DFR2 promoter generated novel stable alleles, w4-dp (dilute purple flowers) and w4-p (pale flowers). We hypothesize that the element is fractured during transposition, and truncated versions of the element in new insertion sites cause stable mutations. The highly active endogenous transposon, Tgm9, should facilitate genomics studies specifically that relate to legume biology.
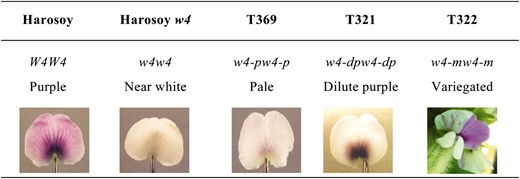
IN soybean [Glycine max (L.) Merr.], five loci W1, W3, W4, Wm, and Wp control the pigmentations in flowers and hypocotyls (Palmer et al. 2004). Soybean plants with genotype W1_ w3w3 W4_ Wm_ Wp_ produce wild-type purple flowers (Figure 1) and purple hypocotyls. Mutations at the W4 locus in the W1_ background result in altered pigment accumulation patterns in petals and reduced levels of purple pigments in flowers and hypocotyls. Four mutant alleles, w4, w4-m, w4-dp, and w4-p have been mapped to this locus. The w4 allele represents a spontaneous mutation, which produces near-white flowers (Figure 1) and green hypocotyls (Hartwig and Hinson 1962; Groose and Palmer 1991). The w4-m allele was identified from a cross between two experimental breeding lines with white and purple flowers, respectively (Palmer et al. 1989; Weigelt et al. 1990). w4-m is characterized by variegated flowers (Figure 1) and green hypocotyls with purple sectors (Groose et al. 1988).
w4-m has been proposed to harbor a class II transposable element (Palmer et al. 1989). Presumably, somatic excision of the putative transposable element results in the variegated (Groose et al. 1988) and germinal excision wild-type phenotypes, purple flowers and purple pigments on hypocotyls (Palmer et al. 1989; Groose et al. 1990). The mutable line carrying w4-m undergoes germinal reversion at a very high frequency, about 6% per generation (Groose et al. 1990). Approximately 1% of the progeny derived from germinal revertants contain new mutations in unlinked loci, presumably resulting from reinsertion of the element (Palmer et al. 1989). For example, female partial-sterile 1 (Fsp1), female partial-sterile 2 (Fsp2), female partial-sterile 3 (Fsp3), and female partial-sterile 4 (Fsp4) were isolated from progenies of germinal revertants with purple flowers and were mapped to molecular linkage groups (MLG) C2, A2, F, and G, respectively (Kato and Palmer 2004). Similarly, 36 male-sterile, female-sterile mutants mapped to the st8 region on MLG J (Kato and Palmer 2003; Palmer et al. 2008a), 24 necrotic root (rn) mutants mapped to the rn locus on MLG G (Palmer et al. 2008b), and three Mdh1-n y20 mutants, mapped to a chromosomal region on MLG H (Palmer et al. 1989; Xu and Palmer 2005b), were isolated among progenies of germinal revertants.
In addition to germinal revertants with purple flowers, the w4 mutable line also generated intermediate stable revertants that produce flowers with variable pigment intensities ranging from purple to near-white (Figure 1). Two stable intermediate revertants, w4-dp and w4-p, are allelic to W4. Plants carrying w4-dp or w4-p alleles produce dilute purple flowers or pale flowers, respectively (Figure 1) (Palmer and Groose 1993; Xu and Palmer 2005a).
Variation in flower color among soybean lines carrying different W4 alleles.
Pigment formation requires two types of genes: structural genes that encode anthocyanin biosynthetic enzymes [e.g., CHS (chalcone synthase), F3H (flavanone 3-hydroxylase), DFR (dihydroflavonol-4-reductase), ANS (anthocyanidin synthase); Figure S1] and regulatory genes that control expression of structural genes (Holton and Cornish 1995). Among the five genes, W1, W3, W4, Wp, and Wm, controlling pigment biosynthesis in soybean, four have been characterized at the molecular level (Figure S1). W1 encodes a flavonoid 5′, 3′-hydroxylase (Zabala and Vodkin 2007). W3 cosegregates with a DFR gene, Wp encodes a flavonone 3-hydroxylase (F3H), and Wm encodes a flavonol synthase (FLS) (Fasoula et al. 1995; Zabala and Vodkin 2005; Takahashi et al. 2007).
Nine CACTA-type class II transposable elements, Tgm1, Tgm2, Tgm3, Tgm4, Tgm5, Tgm6, Tgm7, Tgm-Express1, and Tgmt*, have been reported in soybean (Rhodes and Vodkin 1988; Zabala and Vodkin 2005, 2008). Tgm-Express1 causes mutation in Wp (Zabala and Vodkin 2005) and Tgmt* (EU190440) in T that encodes a flavonoid 3′ hydroxylase (F3′H) (Zabala and Vodkin 2003, 2008). The objectives of the present study were to characterize the W4 locus and then investigate whether the w4-m allele harbors an active transposable element. Our results showed that a CACTA-like transposable element located in a dihydroflavonol-4-reductase gene causes variegated flower phenotype in soybean.
MATERIALS AND METHODS
Primers and probes:
All the primers and probes used in this study are listed in supporting information, Table S1 and Table S2, respectively.
Plant materials:
Soybean lines differing for W4 alleles were planted at the Bruner Farm, the United States Department of Agriculture (USDA) greenhouse or growth cabinet, Iowa State University (Ames, IA). Their genotypes and phenotypes are described in Table 1. For analyses of anthocyanins, flavonols, and RNAs, petals were collected from floral buds 1 day before anthesis. For DNA analyses, genomic DNA was extracted from young leaves.
Soybean lines used in this study
Soybean lines . | Genotypes . | Flower color . | Description of the W4 locus . |
|---|---|---|---|
| Harosoy | W1W1 w3w3 WmWm WpWp W4W4 | Purple | Wild-type |
| T321 | W1W1 w3w3 WmWm WpWp w4-dpw4-dp | Dilute purple | Mutant revertant from T322 (w4-m) |
| T322 | W1W1 w3w3 WmWm WpWp w4-mw4-m | Variegated | Mutable allele |
| T369 | W1W1 w3w3 WmWm WpWp w4-pw4-p | Pale | Mutant revertant from T322 (w4-m) |
| T325 | W1W1 w3w3 WmWm WpWp W4W4 | Purple | Wild-type revertant from T322 (w4-m) |
| Williams | w1w1 w3w3 WmWm WpWp W4W4 | White | Wild-type |
| Minsoy | W1W1 w3w3 WmWm WpWp W4W4 | Purple | Wild-type |
| Williams 82 | w1w1 w3w3 WmWm WpWp W4W4 | White | Wild-type |
Soybean lines . | Genotypes . | Flower color . | Description of the W4 locus . |
|---|---|---|---|
| Harosoy | W1W1 w3w3 WmWm WpWp W4W4 | Purple | Wild-type |
| T321 | W1W1 w3w3 WmWm WpWp w4-dpw4-dp | Dilute purple | Mutant revertant from T322 (w4-m) |
| T322 | W1W1 w3w3 WmWm WpWp w4-mw4-m | Variegated | Mutable allele |
| T369 | W1W1 w3w3 WmWm WpWp w4-pw4-p | Pale | Mutant revertant from T322 (w4-m) |
| T325 | W1W1 w3w3 WmWm WpWp W4W4 | Purple | Wild-type revertant from T322 (w4-m) |
| Williams | w1w1 w3w3 WmWm WpWp W4W4 | White | Wild-type |
| Minsoy | W1W1 w3w3 WmWm WpWp W4W4 | Purple | Wild-type |
| Williams 82 | w1w1 w3w3 WmWm WpWp W4W4 | White | Wild-type |
Soybean lines used in this study
Soybean lines . | Genotypes . | Flower color . | Description of the W4 locus . |
|---|---|---|---|
| Harosoy | W1W1 w3w3 WmWm WpWp W4W4 | Purple | Wild-type |
| T321 | W1W1 w3w3 WmWm WpWp w4-dpw4-dp | Dilute purple | Mutant revertant from T322 (w4-m) |
| T322 | W1W1 w3w3 WmWm WpWp w4-mw4-m | Variegated | Mutable allele |
| T369 | W1W1 w3w3 WmWm WpWp w4-pw4-p | Pale | Mutant revertant from T322 (w4-m) |
| T325 | W1W1 w3w3 WmWm WpWp W4W4 | Purple | Wild-type revertant from T322 (w4-m) |
| Williams | w1w1 w3w3 WmWm WpWp W4W4 | White | Wild-type |
| Minsoy | W1W1 w3w3 WmWm WpWp W4W4 | Purple | Wild-type |
| Williams 82 | w1w1 w3w3 WmWm WpWp W4W4 | White | Wild-type |
Soybean lines . | Genotypes . | Flower color . | Description of the W4 locus . |
|---|---|---|---|
| Harosoy | W1W1 w3w3 WmWm WpWp W4W4 | Purple | Wild-type |
| T321 | W1W1 w3w3 WmWm WpWp w4-dpw4-dp | Dilute purple | Mutant revertant from T322 (w4-m) |
| T322 | W1W1 w3w3 WmWm WpWp w4-mw4-m | Variegated | Mutable allele |
| T369 | W1W1 w3w3 WmWm WpWp w4-pw4-p | Pale | Mutant revertant from T322 (w4-m) |
| T325 | W1W1 w3w3 WmWm WpWp W4W4 | Purple | Wild-type revertant from T322 (w4-m) |
| Williams | w1w1 w3w3 WmWm WpWp W4W4 | White | Wild-type |
| Minsoy | W1W1 w3w3 WmWm WpWp W4W4 | Purple | Wild-type |
| Williams 82 | w1w1 w3w3 WmWm WpWp W4W4 | White | Wild-type |
Extraction and analysis of anthocyanins:
To extract anthocyanin pigments, freeze-dried flower petals were incubated in 1% (v/v) HCl in methanol for 3 hr at room temperature and centrifuged at 13,000 rpm for 10 min. Half of the supernatants was used for spectrophotometric analysis in a Beckman DU 640 nucleic acid and protein analyzer. The other half was hydrolyzed by boiling for 30 min. Hydrolyzed extracts were subjected to spectrophotometric analyses. The anthocyanidin contents were expressed as the absorbance at 535 nm (A535) per milligram of dried petals per milliliter of solvent.
High performance liquid chromatography (HPLC) analysis of flavonols:
The flavonol aglycone samples of soybean flowers and authentic standard solutions of myricetin, quercetin, and kaempferol (Sigma, St. Louis, MO) were prepared according to Burbulis et al. (1996), and stored at −20°. Samples (100 μl) were injected into a C-18 RP column attached to a Waters gradient HPLC system (Millipore, Billerica, MA) and eluted at a flow rate of 1.0 ml/min using the following linear gradient of HPLC-grade acetonitrile in HPLC-grade H2O (pH 3.0, adjusted with glacial acetic acid): 0 to 0% for 5 min, 0 to 10% for 5 min, 10 to 30% for 60 min, 30 to 100% for 5 min, 100 to 100% for 2 min, 100 to 0%, for 2 min, and 0 to 0% for 5 min. The system was run and data were acquired using Waters Millennium software, version 3.2. Elutents were analyzed by a photodiode array 996 detector (PDA996) at 255 nm and quantified by comparing to authentic standards.
RNA preparation, RT–PCR, and RNA blot analysis:
Total RNA was prepared from immature petals using RNeasy mini kit (QIAGEN, Valencia, CA). cDNAs were synthesized from 2 μg total RNAs using oligo-dT and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) and diluted twofold for PCR. Primers for PCR are listed in Table S1. For RNA blot analyses, 20 μg total RNAs was separated on a 1.0% formaldehyde-agarose gel and blotted onto a Zeta Probe Nylon membrane (BioRad, Hercules, CA) by capillary transfer.
DNA preparation and DNA blot analysis:
Genomic DNA was extracted from young leaves by following the CTAB method (Keim et al. 1988), purified with equal volumes of phenol, phenol/chloroform (1:1/v:v), and chloroform (Sambrook et al. 1989). For DNA blot analysis, 10 μg genomic DNA was digested with desired restriction enzymes and separated on a 0.8% (w/v) agarose gel. DNA blot analysis was conducted as previously described (Sambrook et al. 1989).
BAC library screening and W4 gene cloning:
A BAC library (Bhattacharyya et al. 2005) was screened using a partial DFR cDNA probe (Table S2). Positive clones were confirmed by DNA blot analysis. Sequence of the full-length DFR2 gene was obtained through primer walking sequencing method. The BAC DNA for sequencing was extracted using the QIAGEN large constructs miniprep kit.
Genomic library construction and screening:
Two genomic libraries were constructed in the Lambda FIXII/XhoI vector (Stratagene, La Jolla, CA) using the DNA prepared from leaves of the T322 line homozygous for w4-m (Palmer et al. 1990). The DNA from the libraries was transferred to 137-mm nitrocellulose disks (Stratagene) (Sambrook et al. 1989). Aproximately 0.4 million plaques of the first library and 1.5 million plaques of the second library were screened with a DFR2 cDNA fragment (Table S2). Positive clones were confirmed by Southern blot analysis, PCR, and sequencing. The lambda DNA for sequencing was extracted using the QIAGEN Lambda Midi kit.
PCR conditions:
PCR reactions were conducted in a 25-μl mixture containing ∼100 ng of genomic DNA or ∼1 ng of plasmid or phage DNA or 2 μl of first strand cDNA, 1× PCR buffer, 2.0 mm MgCl2, 100 μm dNTP, 0.15 μm of each primer, and 1 unit of Biolase Taq polymerase (Bioline USA, Randolph, MA). PCR was started with an initial 2-min denaturation step at 94° followed by 5 cycles of 94° (30 sec), 60° (1 min, reduced by −1°/cycle), and 72° (1.5 min), and then by 27 cycles of 94° (30 sec), 54° (30 sec), and 72° (30 sec), with a final extension at 72° for 10 min.
DNA sequencing and sequence analysis:
All the sequencing projects were conducted in an ABI 3730 DNA analyzer at the Iowa State University DNA facility. The local alignments were performed using BLAST (bl2seq) from NCBI (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi). The global alignments and multiple alignments were conducted using ClustalW2 from EBI (www.ebi.ac.uk/clustalw2). Gene prediction was performed with GENSCAN (http://genes.mit.edu/GENSCAN.html). Polypeptide sequences were deduced from the DNA sequence using ExPASy translate tool (http://ca.expasy.org/tools/dna.html). Conserved domains in protein were searched with CDS program of NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Accession numbers:
Sequence data can be found in GenBank/EMBL database with accession nos. DQ026299 (DFR2 partial cDNA), EF187612 (DFR2 genomic sequence), EU068464 (truncated Tgm9), EU068463 (insertion in w4-dp), and GQ344503 (Tgm9).
RESULTS
The w4 mutation blocks conversion of dihydromyricetin to delphinidin-3-monoglucoside:
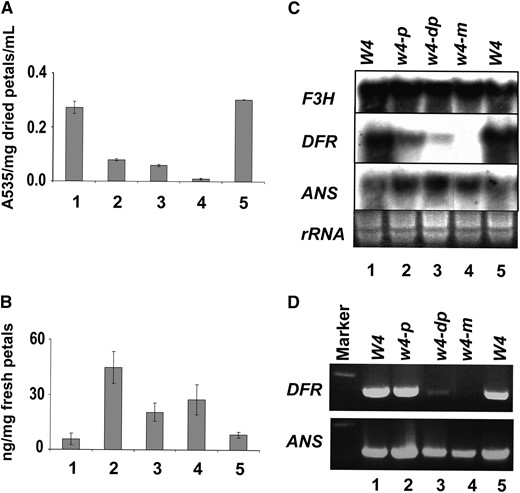
The anthocyanins and flavonols present in flowers of four soybean lines, Harosoy (W4), T322 (w4-m), T321 (w4-dp), and T369 (w4-p), were investigated (Table 1). Anthocyanin extracts showed the maximum absorption peak at 535 nm with λmax 450–650 nm (Figure S2). The peak shifted to 543 nm when the extracts were hydrolyzed by boiling (Figure S2). These spectral characteristics suggested that the main pigment in soybean flowers could be delphinidin-3-monoglucoside or its derivatives, petunidin-3-monoglucoside, and malvidin-3-monoglucoside (Harborne 1958), similar to the main pigment malvidin in soybean hypocotyls, stem, and subepidermal tissues (Nozzolillo 1973). Malvidin is generated through glycosylation and methylation of delphinidin. The anthocyanin contents in flower petal samples were investigated at 535 nm. The highest anthocyanin level was observed in wild-type purple petals (Harosoy) and purple petal sectors of T322, followed by pale flowers (T369) and dilute purple flowers (T321). The lowest anthocyanin content was observed in white petal sectors of T322 (Figure 2A).
Biochemical and molecular analyses of lines carrying individual w4 alleles. (A) Contents of total anthocyanins in five immature petal samples from four soybean lines: 1, Harosoy (purple; W4); 2, T369 (pale; w4-p); 3, T321 (dilute purple; w4-dp); 4, T322w (white sectors of variegated petals; w4-m); and 5, T322p (purple section of variegated petals; W4). (B) Myricetin contents in five petal samples. (C) The transcript level of DFR, ANS, and F3H in five petal samples determined by Northern blot analysis. (D) RT–PCR products of DFR and ANS genes in five petal samples.
Delphinidin-3-monoglucoside or its derivatives are believed to be the main pigments in soybean flowers. The flavonol myricetin is synthesized from a precursor of delphinidin-3-monoglucoside, dihydromyricetin by the enzyme flavonol synthase (FLS; Figure S1). HPLC analyses revealed enhanced accumulation of myricetin in petals of T321 and T369, and white petal sectors of T322 (Figure 2B) that showed less anthocyanin pigment accumulation (Figure 2B). These results suggested that the lesion in w4 mutants is from dihydromyricetin to delphinidin-3-monoglucoside (Figure S1).
Mutations in the W4 locus were associated with reduced DFR2 transcript levels:
We analyzed the w4 mutants for steady state transcript levels of three structural genes, F3H, DFR, and ANS (Figure S1). The probes for F3H and ANS were a cDNA fragment (BM093886) and an RT–PCR product (Table S1 and Table S2), respectively. Results showed that steady state transcript levels of F3H and ANS were comparable among the soybean lines (Figure 2C). The probe for DFR was an RT–PCR product generated from immature flowers using primers DFR1F and DFR1R (Table S1 and Table S2). It encoded a protein named DFR2 that showed 81% amino acid identity to DFR1 (AF167556). The steady state DFR2 transcript level was highest in wild-type Harosoy (W4) and the purple petal sectors of T322, reduced in T369 (w4-p) and T321 (w4-dp), and undetectable in the white petal sectors of T322 (Figure 2C). Similar results for steady state DFR2 transcript levels were observed in RT–PCR analyses (Figure 2D). These data suggested that reduced levels of anthocyanin pigments in w4 mutants (Figure 2A) were the results of lower DFR2 expression levels.
Characterization of T322 (w4-m) and its revertants suggested that DFR2 is located in the W4 locus:
DFR2 was isolated from a BAC library (Bhattacharyya et al. 2005). BAC GS_60E6 was selected for sequencing the entire DFR2 gene (EF187612), which contains six exons and five introns (Figure 3A). The DFR2 coding sequence predicted by GENSCAN (http://genes.mit.edu/GENSCAN.html) was 1065 bp long. The deduced DFR2 polypeptide contained 354 amino acids with 82% identity to DFR1 (AF167556) (Figure S3).
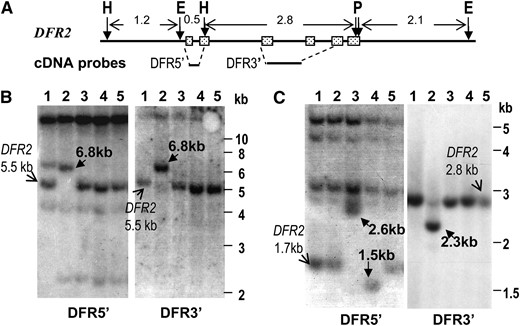
Organization of DFR2 among soybean lines carrying different W4 alleles. (A) Schematic representation of the wild-type DFR2 gene isolated from the BAC clone, GS_60E6. E, EcoRI; H, HindIII; P, PstI. Approximate positions of the probes used in B and C are shown below the DFR2 gene. (B) Organization of EcoRI digested DNA of five soybean lines. Probes are shown below each DNA blot. Slim arrows point to the expected restriction fragments from the wild-type DFR2 gene. Boldface arrows point to the polymorphic fragments in w4 mutants. Lanes 1, Williams (W4); 2, T322 (w4-m); 3, T321 (w4-dp); 4, T369 (w4-p); and 5, T325 (W4). (C) RFLP analyses of HindIII–PstI digested DNA of five soybean lines.
To determine whether DFR2 is the W4 gene and whether insertion of an active, class II transposable element in DFR2 gave rise to the w4-m allele, cultivar Williams (W4), T322 (w4-m), and revertants of T322 [T321 (w4-dp), T369 (w4-p), and T325 (W4)] (Table 1) were studied for organization of DFR2. DFR2 contains a HindIII restriction site in exon II, which divides the DFR2 gene into two halves, 5′ and 3′ ends (Figure 3A). DFR5′ and DFR3′ cDNA probes hybridizing these 5′ and 3′ ends were prepared and used in Southern blot analyses (Figure 3A; Table S2). As expected, the ∼5.5-kb EcoRI fragment was detected by both probes in Williams (W4), T321 (w4-dp), T369 (w4-p), and T325 (W4); but in T322 (w4-m), the fragment was ∼6.8 kb (Figure 3B), suggesting the presence of an insertion in the w4-m allele. Most likely this insertion was excised in the germinal revertants T321 (w4-dp), T369 (w4-p), and T325 (W4).
In HindIII–PstI double digested DNA, probe DFR5′ detected a 1.7-kb HindIII fragment in Williams (W4), T322 (w4-m), and T325 (W4) as expected (Figure 3A), but an ∼2.6-kb fragment in T321 (w4-dp) and an ∼l.5-kb fragment in T369 (w4-p), respectively (Figure 3C, left panel). Probe DFR3′ detected an ∼2.8-kb fragment in Williams (W4), T321 (w4-dp), T369 (w4-p), and T325 (W4), but an ∼2.3-kb fragment in T322 (w4-m) (Figure 3C, right panel). These results suggested that in T322, the insertion is located in the HindIII–PstI DFR2 fragment.
In DFR2, the ∼1.7-kb HindIII fragment includes the promoter, exon I, a part of exon II, and an EcoRI site (Figure 3A). Since no DFR2-specific polymorphisms for EcoRI-digested DNA was observed among wild-type and T321 (w4-dp) or T369 (w4-p) lines (Figure 3B), the aberrations in these two mutants should reside in the ∼1.2-kb HindIII–EcoRI fragment containing the upstream promoter (Figure 3A). These results showed that dfr2 mutations were generated from insertions among the w4 alleles, and therefore W4 most likely encodes DFR2 (Figure 3, B and C).
The insertion in DFR2 intron II is a CACTA-like element Tgm9:
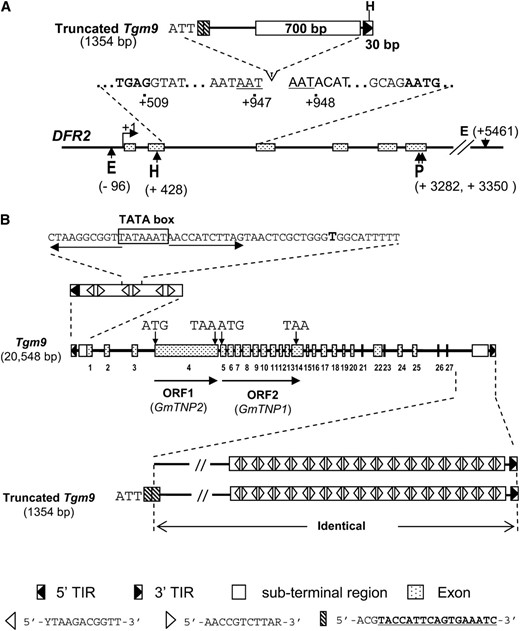
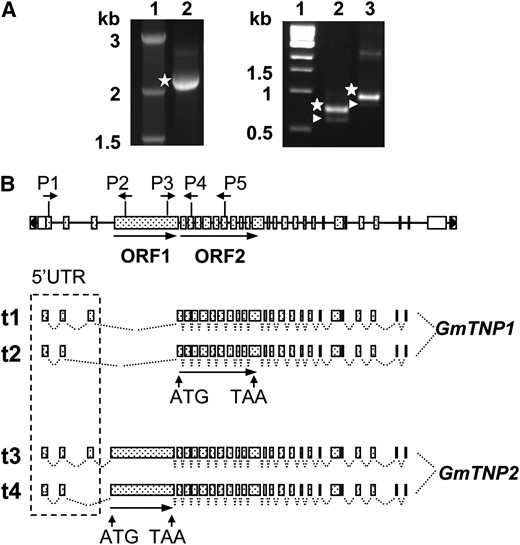
Southern analyses suggested that an insertion was located between DFR2 exon II and VI in T322 (w4-m) (Figure 3). We isolated a 1357-bp insertion in DFR2 intron II, 438 bp downstream of the exon II/intron II junction (Figure 4A). The insertion harbors a HindIII site at the 3′ end, which led to generation of an ∼2.3-kb HindIII–PstI fragment for the w4-m allele when the DNA blot was hybridized with the DFR3′ probe (Figure 3C).
Molecular characterization of the Tgm9 element. (A) Schematic representation of the truncated Tgm9 isolated from the w4-m allele. The positions of nucleotide and restriction sites relative to the transcription start site (TSS) (+1) in wild-type DFR2 are shown. Nucleotides in boldface type were from exons. E, EcoRI; H, HindIII; and P, PstI. (B) Schematic representation of the Tgm9 element. Tgm9 contains a 5′-TIR, 3′-TIR, and 27 exons (marked from 1 to 27) and two ORFs. ORF1 encodes putative transposase GmTNP2. ORF2 encodes putative transposase GmTNP1. A tail-to-tail dimer consisting of two inverted 11-bp motifs repeated 15 times at the 3′ subterminal region (STR) and 3 times at the 5′-STR. A putative TATA box was identified from the second dimer of the 5′ end STR. Truncated Tgm9 element is identical to the 3′ end of the full length Tgm9 element with the exception of the 26 nucleotides (5′-ATTACGTACCATTCAGTGAAATCACG-3′) in its 5′ end that are absent in Tgm9. As a result, two 20-bp (ACGTACCATTCAGTGAAATC) tandem repeats (box filled in with slashes) were identified from the 5′ end of the truncated element.
The inserted element generated a 3-bp (AAT) target site duplication (TSD), similar to TSD generated by CACTA-type transposons (Pereira et al. 1986; Rhodes and Vodkin 1988; Nacken et al. 1991; Inagaki et al. 1994) and contained structures similar to the 3′ end of the CACTA elements. It carried a 30-bp terminal inverted repeat (TIR) starting with 5′-CACTA-3′ similar to the ones in other soybean Tgm elements (Table S4) and a 700-bp highly repetitive region in the subterminal repeat (STR) region next to 3′-TIR (Figure 4). It does not contain other structures such as 5′ end TIR and transposase gene(s), suggesting that it is a truncated version of a transposable element, most likely generated from an imperfect excision of the entire element. We named the entire element Tgm9.
To clone the entire Tgm9, we constructed and screened a genomic library carrying ∼20 genome equivalents DNA prepared from T322 that showed high levels of both somatic excision and germinal reversion. Two nonoverlapping plaques, 16 and 25 carrying 5′ and 3′ ends of Tgm9, respectively, were sequenced (Figure S4 A). By conducting a long range PCR and then a sub PCR (Figure S4 B), a 19-bp (GTTTTGTTGATCATTTACA) missing Tgm9 sequence between the two adjacent ends of clones 16 and 25 was obtained (Figure S4 A). Tgm9 was 20,548 bp (GQ344503). It contained 5′- and 3′-TIR starting with 5′-CACTA-3′, and transposase genes (Figure 4B).
The truncated Tgm9 element was identical to the 3′ end of Tgm9 except for a novel 26-nt sequence (5′-ATTACGTACCATTCAGTGAAATCACG-3′), which with its downstream 17-nt sequence (5′-TACCATTCAGTGAAATC-3′) formed two 20-bp tandem direct repeats (5′-ACGTACCATTCAGTGAAATC-3′) at the 5′ end of the truncated element (Figure 4B). We were able to PCR amplify the truncated element from T322. Therefore, the truncated element most likely arose from imprecise excision of the element. The novel 26-nt sequence was presumably generated through slipped mispairing accompanied by intragenic recombination and deletion as has been documented for generation of a direct repeat (Tavassoli et al. 1999).
Tgm9 showed high sequence identity to Tgmt* isolated recently from the soybean t* allele (Zabala and Vodkin 2008). Only 19 mismatches and 7 half mismatches (6 Rs and 1Y in Tgmt*) were found between the two elements (Table S3). The element was defined by an imperfectly inverted repeat starting with 5′-CACTA-3′. The 3′-STR end was highly structured and contained 12 stem-loop structures, each with a 7-bp motif (5′-AACCGTC-3′) (Zabala and Vodkin 2008). We observed that this 7-bp motif was located within a conserved 11-bp motif (5′-AACCGTCTTAR-3′). This conserved (80%) motif repeated 30 times as 15 tail-to-tail dimers in the 3′-STR region and 6 times as 3 tail-to-tail dimers in the 5′-STR region (Figure 4B).
Alternate splicing generated transposase transcripts in Tgm9:
Zabala and Vodkin (2008) identified 24 exons from the Tgmt* element. All these exons were found in Tgm9 (Figure 4B, exons VI–XXVII) and their expression was detected by RT–PCR in T322. The exons contained two open reading frames (ORF), ORF1 and ORF2 (Figure 4B). By conducting rapid amplification of 5′ complementary DNA ends (5′-RACE), we were able to identify three additional exons (exons I, II, and III) at the 5′ end of the transcripts (Figure 4B). RT–PCR experiments revealed four types of transposase transcripts, t1–t4 (Figure 5).
Expression and alternative splicing of transposase genes in Tgm9. (A) Expression of transposase genes detected in T322 petals by RT–PCR. Twenty-seven exons of Tgm9 were amplified in several RT–PCR experiments as segments and sequenced. For example, cDNA fragment (marked as star) amplified by primers P3 and P5 is shown in lane 2 of the left panel. It covered the region from ORF1 to ORF2. The 5′-UTRs of ORF1 and ORF2 were amplified with primers P1 and P2 (lane 2) or P1 and P4 (lane 3), respectively and are shown in the right panel. Positions of the primers are shown in B. Each primer combination produced two products. Products containing exon III were marked with stars or those without were with arrowheads. Lane 1, marker. (B) Schematic representation of GmTNP1 and GmTNP2 transcripts produced by alternative splicing. Four types of transcripts (t1–t4) with or without exon III or IV were detected. Transcripts t1 and t2 carrying no exon IV, amplified by primers P1 and P4 in A, encode GmTNP1; and t3 and t4 carrying exon IV, amplified by primers P1 and P2 in A, encode GmTNP2. Two ORFs and positions of their start/stop codons and 5′-UTR are shown.
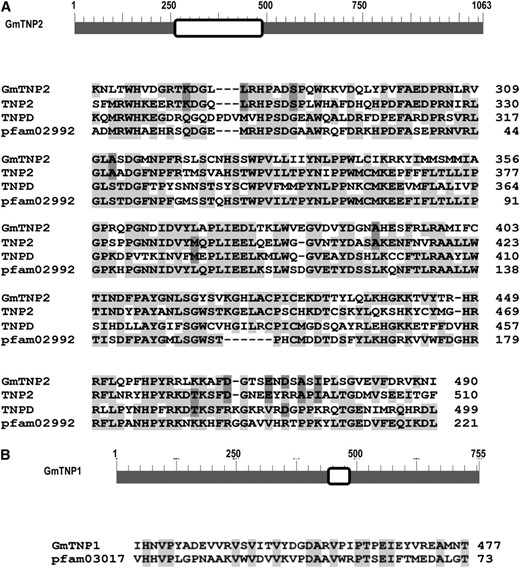
The t1 and t2 transcripts contain ORF2 (Figures 4B and 5B). The 5′ ends of these two transcripts were detected with a forward primer (P1) from exon I and a reverse primer (P4) from exon VI. The 5′ end of t1 contained exons I, II, III, V, and VI, and that of t2 included exons I, II, V, and VI (Figure 5, A and B). The 5′-UTR and ORF2 were identified in exons I–III and exons V–XIV, respectively. Exon IV containing ORF1 was spliced out in both t1 and t2. ORF2, starting at nt 9455 (exon V) and stopping at nt 12,546 (exon XIV), encoded a 755-aa polypeptide containing pfam03017 domain, which belonged to TNP1-like transposase 23 (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Figure 6B). The deduced polypeptide was named GmTNP1. The N terminus of GmTNP1 shared 24% identity with transposase TNP1 in Antirrhinum majus Tam1 (Nacken et al. 1991) but no similarity with transposase TNPA in maize En/Spm (Pereira et al. 1986; Masson et al. 1989).
Conserved domains in GmTNP1 and GmTNP2. (A) Schematic representation of GmTNP2. The conserved domain is boxed. The conserved sequences of the domain among GmTNP2, Tam1 TNP2, En/Spm TNPD, and TNP2_like domain pfam02992 (transposase_21) are shown. (B) Schematic representation of GmTNP1. The conserved domain is boxed. The conserved sequences of the domain between GmTNP1 and TNP1-like domain pfam03017 super family (transposase_23, TNP1/En/Spm) are shown. Conserved amino acids among different proteins are shaded.
The 5′ ends of t3 and t4 transcripts contained exons I–IV and exons I, II, and IV, respectively (Figure 5). The first three exons constitute the 5′-UTR of ORF1. ORF1, starting at nt 6127 and stopping at nt 9316 (exon IV) (Figure 4B), encoded a 1063-aa polypeptide with a conserved domain, pfam02992 (transposase_21, transposase familyTNP2) found in TNP2 of Tam1 (Nacken et al. 1991) and TNPD of En/Spm (Pereira et al. 1986; Masson et al. 1989) (Figure 6A). The deduced polypeptide was named GmTNP2, which shared 32 and 46% identities with TNP2 and TNPD, respectively.
W4 encodes DFR2:
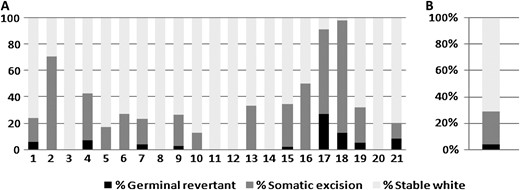
To determine whether the variegated flower phenotype is caused by excision of Tgm9 from DFR2, we investigated >320 progenies of 21 families descended from a single T322 progenitor for hypocotyls and flower colors in greenhouse (Figure 7A). Nine families carried at least some progenies that were either germinal (purple hypocotyls and flowers) or somatic revertants (variegated flowers and purple sectors on hypocotyls). Six other families produced at least some progenies that showed somatic excisions. The average rates of germinal reversion and somatic excisions were 4 and 25%, respectively (Figure 7B), which were comparable to earlier estimates (Groose et al. 1990). A larger proportion (>70%) of the progenies had only white flowers. Imprecise excision of Tgm9 leading to truncated element (Figure 4B) in the target site could be one of the reasons for generation of high proportions of progenies with white flowers (Figure 7A).
Tgm9 is a highly active endogenous transposable element. (A) Investigation of the rates (%) of germinal reversion and somatic excision for flower colors in 21 families generated from a single T322 plant. Germinal reversion produced only purple flowers. Somatic excision produced variegated flowers. Stable white plants produce only white flowers. (B) Percentages of progenies showing germinal reversion with only purple flowers, somatic reversion showing variegated petals and only stable white flowers among all 21 families shown in A.
We sequenced Tgm9 insertion sites of independent germinal revertants with purple flowers and observed distinct footprints among the independent germinal revertants (Figure S5). These results confirmed that excision of Tgm9 from the DFR2 intron II resulted in the expression of DFR2, and thereby, gain of purple flower phenotype. Therefore, W4 encodes DFR2 and somatic excision of the element results in variegated flower phenotype.
w4-dp and w4-p alleles were generated from reinsertion of Tgm9 into the DFR2 promoter:
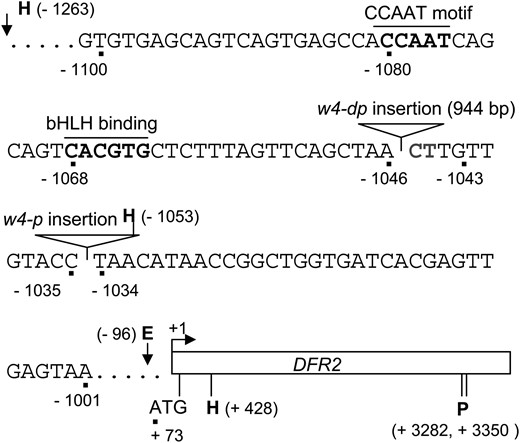
T321 (w4-dp) and T369 (w4-p) mutants were descended from T322 (w4-m). Sequencing of the Tgm9 insertion site confirmed that Tgm9 was excised from DFR2 in both mutants and left behind 4- and 0 (precise excision)-bp footprints in T321 and T369, respectively (Figure S5). The 944-bp insertion (EU068463) in T321 was amplified using primers DFR4S and DFR4R (Table S1). It is identical to the 5′ end of Tgm9. Two nucleotides (C and T) at the 3′ end of insertion site (−1044) were deleted (Figure 8). We failed to PCR amplify the entire insertion in T369. Its 3′ end (381 bp), PCR amplified with primers Tn3′1S and DFR4R (Table S1), was identical to the 3′ end of Tgm9 and located upstream of the −1034th nt of the DFR2 promoter.
Characterization of the w4-dp and w4-p alleles arisen following excision of Tgm9 from the DFR2 intron II. The positions of nucleotide or restriction sites relative to TSS (+1) are shown. Solid triangles indicate the location of insertions in w4-dp and w4-p alleles. E, EcoRI; H, HindIII; and P, PstI.
The insertion sites in the w4-dp and w4-p alleles were only 9 bp apart (Figure 8). The promoter regions between the insertion sites and the transcription start site (TSS) were PCR amplified and sequenced from T321 (w4-dp), T369 (w4-p), and T322 (w4-m). No rearrangements in this region occurred in the mutants (Figure S6). Therefore, the region upstream of Tgm9 insertion sites is important for full expression of DFR2. The upstream promoter regions of structural anthocyanin biosynthesis genes contained cis regulatory elements that affect pigmentation patterns or intensity (Coen et al. 1986; Almeida et al. 1989; Lister et al. 1993). Putative cis-regulatory elements CCAAT motif (Gelinas et al. 1985) and E-box (CACGTG) (Ephrussi et al. 1985) are located upstream of Tgm9 insertion sites in T321 (w4-dp) and T369 (w4-p) (Figure 8), which were moved away from the TSS in both mutants, presumably resulting in reduced expression of DFR2 (Figure 2C).
Tgm9 is a low copy number element:
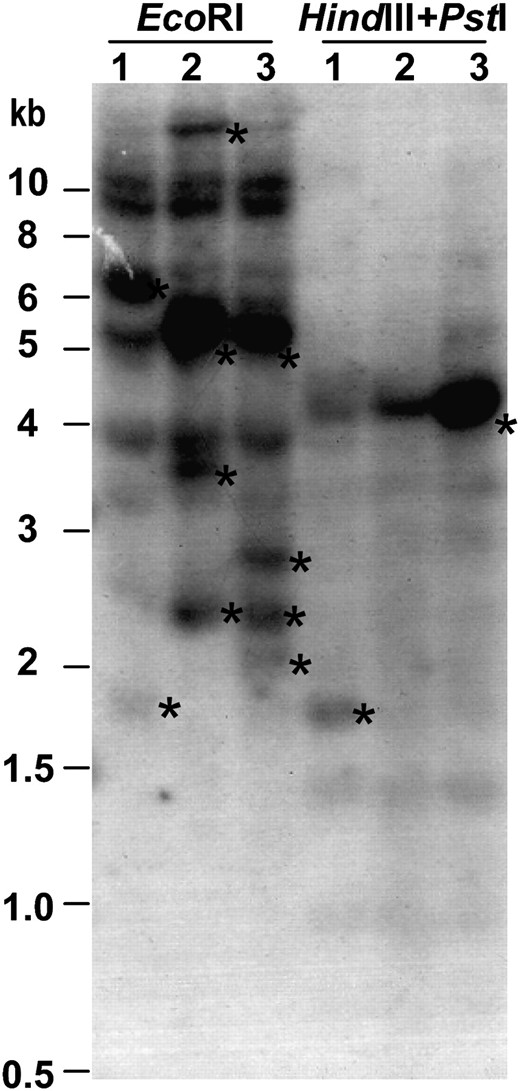
CACTA elements usually have relatively low copy numbers (<100 copies) (Kunze et al. 1997). An earlier study showed that the soybean genome contained 30–42 copies of the Tgm-like elements (Rhodes and Vodkin 1988). The genomic DNA from three NILs, T322 (w4-m), T321 (w4-dp), and T325 (W4), were digested with EcoRI or double digested with HindIII and PstI, and DNA blots were hybridized to the 3′ end of Tgm9. More than 10 copies of the Tgm9-like sequences were detected (Figure 9). T325 was isolated as a germinal revertant with purple flowers from T322. HindIII and PstI digested DNA showed the excision of Tgm9 from the DFR2 intron II and reinsertion into a new locus (Figure 9).
Organization of Tgm9 among soybean lines that vary for the W4 alleles. The probe is the 3′ end of Tgm9. Soybean lines, their W4 alleles, and restriction enzymes used were labeled above individual lanes. Lanes 1, T322 (w4-m); 2, T321 (w4-dp); and 3, T325 (W4). T321 and T325 were isolated as intermediate or full revertant lines, respectively, from T322. Polymorphic bands are shown with *. The EcoRI-specific polymorphisms among three lines were arisen most likely due to cytosine methylation in some of the EcoRI sites. The strong ∼4.5-kb HindIII–PstI band in T325 indicated additional copy resulting from reinsertion of Tgm9 into a new locus.
The recently available soybean genome sequence (http://www.phytozome.org) was searched for Tgm9 5′ end (400 bp), 3′ end (700 bp), GmTNP1, and GmTNP2 sequences. The 5′ end showed similarities to 32 sequences. The 3′ end and GmTNP1 showed similarities to ∼100 sequences of the soybean genome. At least 1500 bp GmTNP2 sequences showed similarity to 1000 sequences of the genome. This suggested that a TNP2-like domain could be conserved among different CACTA elements such as Tgm5 (Rhodes and Vodkin 1988) or functionally related distant proteins. Among the Tgm9-like sequences, one localized to scafold_57 from nt 95,650 to 13,598 is 99% identical to Tgm9. We named this sequence Tgm10. Compared to Tgm9, Tgm10 is truncated for the first ∼4100-bp sequence, contains a gap in its 5′ end, and a 1049-bp insert in exon XXIII (Figure S7). Tgmt*, Tgm 9, and Tgm10 could be variants from a progenitor element or alternatively, highly active Tgm9 could be a progenitor of Tgmt* and Tgm10.
DISCUSSION
In soybean, the w4-m allele regulates variegated flower color in petals and purple sectors on stems or hypocotyls. By applying biochemical and molecular approaches, we have established that somatic excision of a CACTA-type transposable element Tgm9 from DFR2 encoding dihydroflavonol-4-reductase results in variegated flowers in mutable T322 line carrying the w4-m allele. Tgm9 is ∼20.5 kb long and a member of the CACTA super family of transposons (Pereira et al. 1986; Rhodes and Vodkin 1988; Nacken et al. 1991; Inagaki et al. 1994). It generates 3-bp target-site duplication upon insertion. Its 5′ and 3′ ends carry imperfect terminal inverted repeats (TIRs) flanking the conserved CACTA sequence. Subterminal regions are highly structured and contain multiple copies of putative transposase binding motif (AACCGTCTTAR) (Figure 4) (Gierl et al. 1988). It excises at a high frequency (Figure 7). Excision of Tgm9 generated 8- to 5-bp footprints (Figure S5), which are comparable to the ones created by other CACTA elements such as petunia PsI (Snowden and Napoli 1998).
The excision mechanism in Tgm9 could be similar to one considered for En/Spm (Gierl et al. 1988, 1989; Frey et al. 1990). Through alternative splicing, Tgm9 produces two distinct transposases, GmTNP1 (755 aa, Tam1 TNP1-like transposase) and GmTNP2 (1063 aa, Tam1 TNP2-like transposase) (Figures 4B, 5, and 6). Organization of GmTNP2 and GmTNP1 is comparable to the one observed for transposases TNPA and TNPD in the maize En/Spm element (Pereira et al. 1986; Masson et al. 1989). GmTNP1 is presumably a DNA-binding protein like TNPA, recognizing and binding to the short repetitive motif of the subterminal regions (Gierl et al. 1988). GmTNP2 most likely is an endonuclease-like TNPD (Gierl et al. 1989; Frey et al. 1990). It binds to GmTNP1, interacts with Tgm9 TIRs, pulls the two ends of the element together to form a loop, and excises the element from its insertion site.
The maize En/Spm element preferentially transposes to linked loci (Peterson 1970; Nowick and Peterson 1981). Similarly, Tgm9 transposed into the DFR2 promoter (Figures 3 and 8). However, with the exception of the mutations induced in the DFR2 promoter (e.g., w4-dp and w4-p) (Figures 3 and 8), mutations identified in Tgm9-tagging experiments were mapped to unlinked loci (Palmer et al. 1989, 2008a,b; Kato and Palmer 2003, 2004; Xu and Palmer 2005b).
Tgm9 showed high identity to the Tgmt* element (EU190440, 20,544 bp) isolated from the soybean t* allele (Zabala and Vodkin 2008) (Table S3). As shown here and earlier, Tgm9 is an active element (Palmer et al. 1989; Groose et al. 1990) (Figure 7); whereas, Tgmt* at the soybean t* allele seems not to be (Zabala and Vodkin 2003, 2008). The transposase genes were silenced in line 37609 (t*) (Zabala and Vodkin 2008). High similarity between Tgmt* and Tgm9 suggested that Tgm9 could be the progenitor element of Tgmt*. Tgmt* is comparable to the cryptic spm element from the maize a-m2-8167B allele that contained an intact spm element with no activity (Masson et al. 1987; Banks et al. 1988).
Like most CACTA elements, Tgm9 is a low copy transposable element (Rhodes and Vodkin 1988; Kunze et al. 1997). Active low copy endogenous transposable elements have been considered useful tools in gene cloning and functional genomics studies (Maes et al. 1999; Walbot 2000; Ramachandran and Sundaresan 2001). We expect that highly active Tgm9 should facilitate functional genomics studies in soybean. Genetic data strongly suggested that mutations such as necrotic root (rn), male-sterility, and female sterility (st8) (Palmer et al. 2008a,b) most likely resulted from insertion of Tgm9. Except for two mutations in fertility genes, no reversions events have been observed among the mutants presumably tagged by Tgm9 (R. Palmer, unpublished data).
Truncated Tgm10 and fractured Tgm9 in w4-dp and w4-m allele (EU068463 and EU068464; Figure S7) suggested existence of fractured Tgm9 elements in the soybean genome. Fractured Ac (fAc) elements have been documented in maize (Ralston et al. 1989; Zhang and Peterson 1999). We hypothesize that the element is frequently fractured during transposition events and truncated Tgm9 derivatives cause stable mutations. If our hypothesis is correct, then the element will be useful in creating stable mutations and cloning soybean genes through Tgm9-tagging experiments. To date, to our knowledge no active, endogenous transposable elements have been cloned from any legume species. Therefore, Tgm9 is expected to expedite the genomics research in soybean, and thereby contribute significantly toward our understanding of the legume biology.
Footnotes
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107904/DC1.
Footnotes
Communicating editor: E. J. Richards
Acknowledgements
The authors thank R. W. Groose, University of Wyoming, Laramie, WY, for providing flower pictures; R. C. Shoemaker, United States Department of Agriculture Agricultural Research Service (USDA ARS) and Iowa State University, Ames, IA, for providing the EST clone Gm-c1086-2103; and M. P. Scott, USDA ARS and Iowa State University, for the guidance on the HPLC analysis. We also thank R. Takahashi, National Institute of Crop Science, Tsukuba, Japan and L. Vodkin, Department of Crop Sciences, University of Illinois, Champaign-Urbana, IL for critically reviewing an earlier version of the manuscript and Cathie Martin, John Innes Center, United Kingdom, for reviewing the manuscript. This is a joint contribution of the Iowa Agriculture and Home Economics Experiment Station, Ames, Iowa, Project No. 4403, and the USDA, Agricultural Research Service, Corn Insects and Crop Genetics Research Unit, and was supported by the Hatch Act and the State of Iowa. The mention of a trademark or proprietary product does not constitute a guarantee or warranty of the product by Iowa State University or the USDA, and the use of the name by Iowa State University or the USDA implies no approval of the product to the exclusion of others that may also be suitable.
References
Almeida, J., R. Carpenter, T. P. Robbins, C. Martin and E. S. Coen,
Banks, J. A., P. Masson and N. Fedoroff,
Bhattacharyya, M. K., N. N. Narayanan, H. Gao, D. K. Santra, S. S. Salimath et al.,
Burbulis, I. E., M. Iacobucci and B. W. Shirley,
Coen, E. S., R. Carpenter and C. Martin,
Ephrussi, A., G. M. Church, S. Tonegawa and W. Gilbert,
Fasoula, D. A., P. A. Stephens, C. D. Nickell and L. O. Vodkin,
Frey, M., J. Reinecke, S. Grant, H. Saedler and A. Gierl,
Gelinas, R., B. Endlich, C. Pfeiffer, M. Yagi and G. Stamatoyannopoulos,
Gierl, A., S. Lutticke and H. Saedler,
Gierl, A., H. Saedler and P. A. Peterson,
Groose, R. W., and R. G. Palmer,
Groose, R. W., H. D. Weigelt and R. G. Palmer,
Groose, R. W., S. M. Schulte and R. G. Palmer,
Harborne, J. B.,
Hartwig, E. E., and K. Hinson,
Holton, T. A., and E. C. Cornish,
Inagaki, Y., Y. Hisatomi, T. Suzuki, K. Kasahara and S. Iida,
Kato, K. K., and R. G. Palmer,
Kato, K. K., and R. G. Palmer,
Keim, P., T. Olson and R. C. Shoemaker,
Kunze, R., H. Saedler and W. E. Lönnig,
Lister, C., D. Jackson and C. Martin,
Maes, T., P. De Keukeleire and T. Gerats,
Masson, P., R. Surosky, J. A. Kingsbury and N. V. Fedoroff,
Masson, P., G. Rutherford, J. A. Banks and N. Fedoroff,
Nacken, W. K., R. Piotrowiak, H. Saedler and H. Sommer,
Nowick, E. M., and P. A. Peterson,
Nozzolillo, C.,
Palmer, R. G., and R. W. Groose,
Palmer, R. G., B. R. Hedges, R. S. Benavente and R. W. Groose,
Palmer, R. G., R. W. Groose, H. D. Weigelt and J. E. Miller,
Palmer, R. G., T. W. Pfeiffer, G. R. Buss and T. C. Kilen,
Palmer, R. G., D. Sandhu, K. Curran and M. K. Bhattacharyya,
Palmer, R. G., L. Zhang, Z. Huang and M. Xu,
Pereira, A., H. Cuypers, A. Gierl, Z. Schwarz-Sommer and H. Saedler,
Peterson, P. A.,
Ralston, E., J. English and H. K. Dooner,
Ramachandran, S., and V. Sundaresan,
Rhodes, P. R., and L. O. Vodkin,
Sambrook, J., E. F. Fritsch and T. Maniatis,
Snowden, K. C., and C. A. Napoli,
Takahashi, R., S. M. Githiri, K. Hatayama, E. G. Dubouzet, N. Shimada et al.,
Tavassoli, K., A. Eigel and J. Horst,
Walbot, V.,
Weigelt, H. D., R. G. Palmer and R. W. Groose,
Xu, M., and R. G. Palmer,
Xu, M., and R. G. Palmer,
Zabala, G., and L. Vodkin,
Zabala, G., and L. Vodkin,
Zabala, G., and L. Vodkin,
Zabala, G., and L. O. Vodkin,
Zhang, J., and T. Peterson,