-
PDF
- Split View
-
Views
-
Cite
Cite
Taulant Bacaj, Yun Lu, Shai Shaham, The Conserved Proteins CHE-12 and DYF-11 Are Required for Sensory Cilium Function in Caenorhabditis elegans, Genetics, Volume 178, Issue 2, 1 February 2008, Pages 989–1002, https://doi.org/10.1534/genetics.107.082453
Close - Share Icon Share
Abstract
Sensory neuron cilia are evolutionarily conserved dendritic appendages that convert environmental stimuli into neuronal activity. Although several cilia components are known, the functions of many remain uncharacterized. Furthermore, the basis of morphological and functional differences between cilia remains largely unexplored. To understand the molecular basis of cilia morphogenesis and function, we studied the Caenorhabditis elegans mutants che-12 and dyf-11. These mutants fail to concentrate lipophilic dyes from their surroundings in sensory neurons and are chemotaxis defective. In che-12 mutants, sensory neuron cilia lack distal segments, while in dyf-11 animals, medial and distal segments are absent. CHE-12 and DYF-11 are conserved ciliary proteins that function cell-autonomously and are continuously required for maintenance of cilium morphology and function. CHE-12, composed primarily of HEAT repeats, may not be part of the intraflagellar transport (IFT) complex and is not required for the localization of some IFT components. DYF-11 undergoes IFT-like movement and may function at an early stage of IFT-B particle assembly. Intriguingly, while DYF-11 is expressed in all C. elegans ciliated neurons, CHE-12 expression is restricted to some amphid sensory neurons, suggesting a specific role in these neurons. Our results provide insight into general and neuron-specific aspects of cilium development and function.
SENSORY cells, such as vertebrate photoreceptors, mechanosensory hair cells in the ear, and olfactory neurons, project nonmotile cilia (Wheatleyet al. 1996). These sensory cilia are positioned at the interface between an animal and its environment and transduce information via cell-surface receptors, signal transduction molecules, and specialized ion channels to the nervous system (Pazour and Witman 2003; Scholey and Anderson 2006; Singla and Reiter 2006). The ability of cilia to act as sensory transduction sites is due, at least in part, to their specialized morphologies, which allow for compartmentalization of sensory and signaling components. How this specialized ciliary architecture arises and how signaling molecules are organized within cilia remain poorly understood, although genomic and proteomic studies have identified sets of proteins conserved in all cilia (Avidor-Reisset al. 2004; Liet al. 2004; Blacqueet al. 2005; Efimenkoet al. 2005).
The hermaphrodite nematode C. elegans contains 60 ciliated sensory neurons, 28 of which are assembled into the bilateral amphid and phasmid sensilla (Wardet al. 1975; Whiteet al. 1986), located in the head and tail of the animal, respectively. C. elegans sensory cilia are comprised of a proximal basal body, a medial segment containing doublet microtubules, and a distal segment consisting of singlet microtubules (Wardet al. 1975; Perkinset al. 1986). Some C. elegans sensory neurons end in modified cilia that have a wing- or finger-like morphology (Wardet al. 1975). Defects in amphid cilia structure result in behavioral deficits, as manifested by abnormal chemotaxis to soluble attractants (Che phenotype) or to volatile attractants and repellants (Perkinset al. 1986; Bargmannet al. 1993). Some amphid sensory neurons can take up and concentrate lipophilic dyes from their surroundings through a pore and channel generated by two associated glial cells (Hedgecocket al. 1985; Perens and Shaham 2005). This dye uptake is generally disrupted in animals with abnormal cilia (Dyf phenotype) (Perkinset al. 1986), providing a convenient assay for identifying mutants with defects in sensory cilia biogenesis and function.
The assembly and maintenance of C. elegans cilia, like that of flagella, depend on intraflagellar transport (IFT), the process by which IFT particles are thought to transport cargo in and out of the cilium (Scholey 2003). Studies of Chlamydomonas reinhardtii flagella suggest that each IFT particle is composed of two biochemically defined complexes, A and B (Rosenbaum and Witman 2002). The anterograde movement of the IFT particle is powered by kinesin-2 molecular motors (Orozcoet al. 1999; Snowet al. 2004), while the retrograde movement depends on an IFT-specific dynein (Signoret al. 1999). In C. elegans, several components of the IFT-A and IFT-B subparticles have been characterized and shown to move within cilia. Disruption of these components results in Che and Dyf defects (Colletet al. 1998; Scholey 2003; Bellet al. 2006; Efimenkoet al. 2006).
Mutants in the C. elegans che-12 and dyf-11 genes were isolated in genetic screens for animals displaying dye-uptake defects (Perkinset al. 1986; Starichet al. 1995). To determine whether these genes encode components of the ciliary proteome, we cloned and characterized them. Each gene encodes a conserved protein that localizes within sensory neuron cilia. CHE-12 is a novel protein that does not appear to be part of the IFT particle itself, but is required for sensory cilia function. DYF-11 is a component of the IFT-B complex and is required at an early step of IFT-B particle assembly. Whereas dyf-11 is expressed in many, if not all, sensory neurons, che-12 seems to be expressed only in a subset of amphid neurons. Furthermore, che-12 mutants show differential defects in amphid functions, suggesting that CHE-12 may provide insight into how morphological and functional differences between cilia arise.
MATERIALS AND METHODS
C. elegans strains were cultured at 20° as described (Brenner 1974), unless otherwise indicated. The alleles and strains used in this work were as follows: N2 Bristol as wild type; CB4856 (Hodgkin and Doniach 1997); LGI, che-13(e1805) and daf-16(mu86); LGII, daf-19(m86); LGV, egl-9(n586), che-12(e1812, mn389, mn399), unc-42(e270), che-11(e1810), and bbs-8(nx77); and LGX, dyf-11(mn392) and osm-5(m184). The following transgenic arrays were used: ntIs1 [gcy-5 pro∷GFP, lin-15(+)] V (Sarafi-Reinachet al. 2001), kyIs136 [str-2 pro∷GFP, lin-15(+)] X (Troemelet al. 1999), yhEx90 [che-13 pro∷che-13∷GFP, rol-6(su1006)] (Haycraftet al. 2003), myEx10 [che-11 pro∷che-11∷GFP, rol-6(su1006)] (Qinet al. 2001), nsEx1441 [che-12 pro∷che-12∷GFP, rol-6(su1006)], nsEx1448 [che-12 pro∷GFP, rol-6(su1006)], nsEx1558 [hsp-16.2 pro∷che-12, rol-6(su1006)], nsEx1561 [sra-6 pro∷che-12, rol-6(su1006)], nsEx1761 [gcy-5 pro∷mCherry∷SL2∷che-12∷GFP, rol-6(su1006)], nsEx1702 [dyf-11 pro∷GFP, rol-6(su1006)], nsEx1705 [dyf-11 pro∷dyf-11∷GFP, rol-6(su1006)], nsEx1862 [hsp-16.2 pro∷dyf-11, rol-6(su1006)], nsEx1859 [sra-6 pro∷dyf-11, rol-6(su1006)], and nsEx1764 [gcy-5 pro∷mCherry∷SL2∷dyf-11∷GFP, rol-6(su1006)].
Dye-uptake and behavioral analysis:
Stock solutions (20 mg/ml) of fluorescein-5-isothiocyanate (FITC “isomer I”) (Molecular Probes, Eugene, OR) and 5 mg/ml 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) (Molecular Probes) in N,N-dimethylformamide were stored at −20°. To assay dye uptake, animals were soaked in 0.4 mg/ml FITC or 5 μg/ml DiI diluted in M9 for up to 4 hr. NaCl chemotaxis and odortaxis assays were performed as previously described (Ward 1973; Bargmannet al. 1993). The ring assay (Culotti and Russell 1978) was used to test osmosensation. Briefly, a 1-cm ring of 4 m fructose containing the dye Congo Red was made on an NGM plate. Animals were placed inside the ring and followed over the next 10 min to determine response to the osmotic barrier. Animals exiting the ring were classified as defective.
Mapping and cloning:
unc-42(e270), che-12(mn399), and egl-9(n586) hermaphrodites were crossed to CB4856 (Hodgkin and Doniach 1997) males, which contain many single nucleotide polymorphisms (SNPs) with respect to N2 (Wickset al. 2001). From the F2 progeny, Egl non-Unc and Unc non-Egl animals were selected. These recombinants were soaked in FITC to determine their che-12 genotype. DNA was prepared from the progeny of recombinant animals as previously described (Wickset al. 2001). SNP analysis limited the genomic location of che-12 between the SNPs C12D8:34312 and AC3:3025. Cosmid clones containing DNA spanning this region were injected singly into che-12 animals and transgenic animals were tested for rescue of the FITC uptake defect. Cosmid B0024 was the only one to give rescue. Shorter regions of B0024 were amplified by PCR and introduced into che-12 mutants. A fragment containing the gene B0024.8 yielded animals rescued for dye filling.
Previous work had established that dyf-11(mn392) was located on the left arm of chromosome X (Starichet al. 1995). To refine this position, dyf-11(mn392) hermaphrodites were crossed to CB4856 males. From this cross, 238 dye-filling defective F2 animals were isolated, and DNA prepared from their progeny was characterized for the presence of N2 and CB4856 SNPs. This analysis revealed that dyf-11 resides between the SNPs F39H12:15494 and F02G3:5645. Sequencing candidate genes from this region revealed that the gene C02H7.1 contained a nonsense mutation in dyf-11 animals. Injection of both the C02H7 cosmid and the C02H7.1 genomic region restored dye filling.
Plasmid constructs:
To make che-12 pro∷GFP, the 936 bp immediately upstream of the B0024.8 start site were amplified by PCR and cloned into the SalI/BamHI restriction sites of the pPD95.75 vector (Milleret al. 1999). The full cDNA of B0024.8 was amplified by PCR from a mixed-stage cDNA library (Schumacheret al. 2005) and ligated into the KpnI/AgeI sites of che-12 pro∷GFP, to make a translational fusion construct. To make dyf-11 pro∷GFP, the 1868 bp immediately upstream of the C02H7.1 start site were PCR amplified and cloned into the SphI/SalI sites of the pPD95.69 vector (Milleret al. 1999). The C02H7.1 cDNA was amplified by PCR from a cDNA library (Schumacheret al. 2005) and cloned into the SalI/BamHI sites of pPD95.75. Into this last vector was inserted an SphI/SalI DNA fragment containing the dyf-11 promoter, to yield dyf-11 pro∷dyf-11∷GFP. To make heat-shock-inducible constructs, the che-12 and dyf-11 cDNAs were inserted into pPD49.78 (Fireet al. 1990) as KpnI/SacI and NgoMIV/XmaI fragments, respectively. The che-12 and dyf-11 cDNAs were inserted downstream of a 2.4-kb promoter region of the gene sra-6 (Troemelet al. 1995) as XmaI/SacI and NgoMIV/XmaI fragments, respectively. To make dual-expression constructs, the che-12 and dyf-11 cDNAs were inserted downstream of the SL2 acceptor sequence (Huanget al. 2001) in the vector gcy-5 pro∷mCherry∷SL2 acceptor site∷GFP.
Germ-line transformations:
To generate transgenic animals, adult hermaphrodites were transformed using standard protocols (Melloet al. 1991). Constructs were injected at 10–60 ng/μl along with the co-injection plasmid pRF4, which contains the dominant marker rol-6(su1006) (Melloet al. 1991). Heterozygous che-12/+ animals were used for injections as it was difficult to obtain transgenic lines by injecting che-12 animals directly.
Heat shock:
Animals were placed at 34° for 30 min, allowed to recover at 20° for 6 hr, and scored for dye uptake as described above.
Microscopy and imaging:
GFP expression patterns were analyzed in stable transgenic lines by conventional fluorescence microscopy using an Axioplan II microscope equipped with an AxioCam camera (Carl Zeiss, Thornwood, NY). Expression patterns for each gene were examined in three independent transgenic lines.
IFT was assayed as previously described (Orozcoet al. 1999; Ouet al. 2005). Transgenic animals anesthetized with 5 mm levamisole were mounted on agar pads and imaged at 21°. Imaging was performed on a Zeiss Axiovert 200M microscope equipped with an UltraView spinning disk confocal head using a 100×/1.45 NA objective. Images were collected with a Hamamatsu EM-CCD (C9100-12) camera at 0.333 sec/frame for 5 min. Kymographs were created using the MetaMorph software (Molecular Devices).
Electron microscopy:
Animals were fixed, stained, embedded in resin, and serially sectioned using standard methods (Lundquistet al. 2001). Imaging was performed with an FEI Tecnai G2 Spirit BioTwin transmission electron microscope equipped with a Gatan 4K × 4K digital camera.
RESULTS
che-12 animals exhibit a restricted set of dye-uptake and behavioral defects:
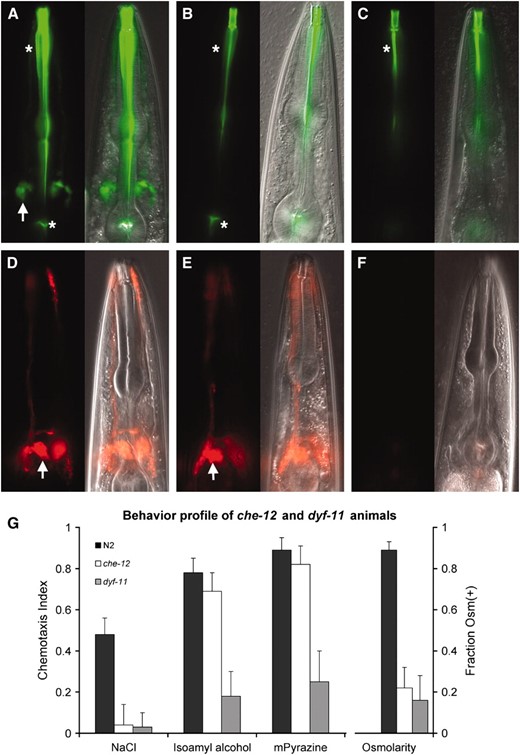
Amphid sensory neurons of C. elegans mutants defective in cilium formation and function fail to concentrate lipophilic dyes, such as FITC and DiI, from their surroundings (e.g., Figure 1, A and D) and fail to perform some sensory neuron-mediated behaviors, such as chemotaxis (Perkinset al. 1986). Three alleles of the che-12 gene, e1812, mn389, and mn399, were previously isolated on the basis of the inability of animals carrying these alleles to concentrate FITC (Perkinset al. 1986; Starichet al. 1995). To characterize mutants homozygous for these alleles in greater depth, we tested them in dye-uptake and behavioral assays. One hundred percent of che-12(e1812), che-12(mn389), and che-12(mn399) animals failed to take up FITC in amphid neurons (n = 100 for each allele; Figure 1B). However, only 2% of che-12(e1812) and che-12(mn389) mutants, and 51% of che-12(mn399) mutants, failed to take up DiI (n = 100 for each allele; Figure 1E), a selectivity that has not been previously described.
Characterization of che-12 and dyf-11 mutants. (A) Uptake of FITC by amphid sensory neurons in a wild-type animal. Left, fluorescence image; right, fluorescence and DIC overlay of the same image. Arrow, cell bodies; asterisks, nonspecific pharyngeal staining. (B) A che-12(mn389) mutant showing FITC-uptake defect. (C) dyf-11(mn392) animals also fail to take up FITC. (D) DiI staining pattern of a wild-type animal. (E) che-12(mn389) animals can also take up DiI. (F) dyf-11(mn392) animals fail to take up DiI. (G) Behavioral defects of che-12(mn399) and dyf-11(mn392) mutants. Both mutants fail to chemotax to 0.2 m NaCl and are unable to respond to an osmotic barrier. dyf-11, but not che-12, animals show defects in odortaxis toward 1% isoamyl alcohol and 1% methyl pyrazine. In all figures anterior is up unless otherwise indicated.
Contrary to a previous report describing normal chemotaxis of che-12(e1812) mutants toward the soluble attractant NaCl (Perkinset al. 1986), we observed defects in this ASE amphid neuron-mediated behavior (Bargmann and Horvitz 1991) in animals homozygous for any of the known che-12 alleles (Figure 1G). Furthermore, all che-12 mutants were defective in avoidance of a high osmolarity 4-m fructose barrier, a behavior mediated in wild-type animals by the ASH amphid neurons (Kaplan and Horvitz 1993) (Figure 1G). However, che-12 mutants were normally attracted to the volatile odorants isoamyl alcohol (1%) and methyl pyrazine (1%; Figure 1G), behaviors mediated by the AWC and AWA amphid neurons (Bargmannet al. 1993), respectively. Thus, che-12 is required for some, but not all, amphid cilia functions.
dyf-11 is required for sensory neuron dye uptake and chemotaxis:
Mutants in the dyf-11 gene were also previously isolated in screens for animals exhibiting dye-uptake defects (Starichet al. 1995), suggesting that this gene may affect cilium structure and/or function. To further characterize the phenotype of dyf-11 mutants, we tested these animals in dye-uptake and behavior assays. A single dyf-11 allele, mn392, is known, and we found that mutants homozygous for this allele displayed a fully penetrant defect in concentrating the two lipophilic dyes FITC and DiI (Figure 1, C and F), consistent with previous studies (Starichet al. 1995). dyf-11(mn392) animals also exhibited strong defects in chemotaxis toward NaCl, isoamyl alcohol, and methyl pyrazine and in avoidance of a high osmolarity barrier (Figure 1G). We further noted that dyf-11 mutants could not enter the dauer state, and mutant males showed low mating efficiency (data not shown), defects that are characteristic of impaired cilia function. Taken together, therefore, these results suggest that dyf-11 is required for normal sensory neuron function and is likely to play a role within cilia.
Cilia of che-12 and dyf-11 mutants are structurally abnormal:
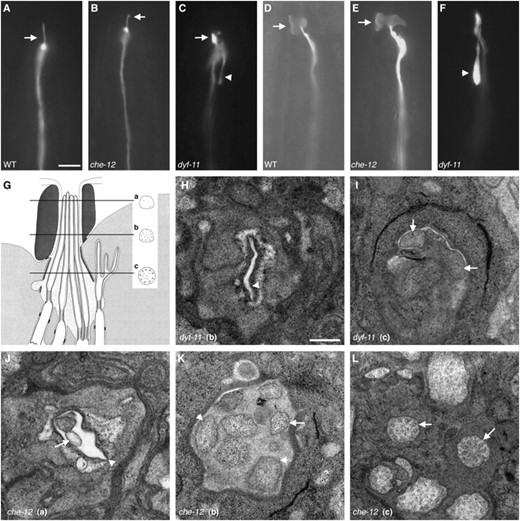
To determine the cause of the Dyf and Che defects of che-12 and dyf-11 mutants, we examined the structures of amphid cilia in these mutants using fluorescence and electron microscopy. We first focused on the ASE neuron, which mediates attraction to NaCl, a behavior defective in both che-12 and dyf-11 mutants. As observed by fluorescence microscopy (Figure 2, A and B), the ASER cilium was shortened in che-12(mn399) animals expressing the gcy-5 pro∷GFP ASER-specific reporter transgene (Yuet al. 1997), with an average cilium length of 4.3 ± 0.5 (n = 10) μm compared to the wild-type average ASER cilium length of 6.1 ± 0.6 μm (n = 10). Cilia were even more severely shortened in dyf-11(mn392) animals expressing the same reporter transgene (average length of 1.4 ± 0.3 μm, n = 10; Figure 2C). These morphological defects could account for the observed defects in ASE-mediated NaCl chemotaxis of che-12 and dyf-11 mutants.
che-12 and dyf-11 mutants have defects in cilium structure. (A) The ASER cilium (arrow) of a wild-type animal expressing gcy-5 pro∷GFP. (B) che-12(mn399) animals have a short ASER cilium. (C) The stunted ASER cilium (arrow) of a dyf-11 animal. A backward process is visible (arrowhead). (D) The AWC cilium (arrow) of a wild-type animal expressing str-2 pro∷GFP. (E) In che-12(mn399), the AWC cilium retains its winged morphology (arrow). (F) The AWC cilium fails to spread in dyf-11 animals. A backward process is present (arrowhead). Bar, A–F, 5 μm. (G) A cartoon of the amphid channel, adapted from Perkinset al. (1986). Three of the eight channel neurons are depicted. The locations of EM cross sections are shown. The normal cross-sectional profile of a cilium at each level is diagrammed. (H) EM cross section at level “b” of a dyf-11(mn392) animal. Note the empty cavity (arrowhead) where amphid cilia should be located. (I) EM cross section of a dyf-11(mn392) animal at level “c” showing the absence of doublet or any other microtubules in neuronal profiles (arrows). (J) EM cross section of a che-12(mn389) mutant at level “a”. The amphid cilia are short, and so only 2 of the 10 neuronal profiles are evident. Amphid opening (arrowhead). The beginning of a cilium (arrow). (K) EM cross section of the distal segment of a che-12(mn389) animal at level “b”. No singlet microtubules are seen in most cilia (arrowheads). A cilium containing doublet microtubules is shown (arrow). (L) Proximal EM cross section of a che-12(mn389) animal at level “c”. The cilia appear normal and contain doublet microtubules (arrows). Bar, H–L, 300 nm. In H–L, dorsal is up.
Examination of che-12(mn389) and dyf-11(mn392) mutants by electron microscopy (EM) revealed defects consistent with the fluorescence imaging studies. Specifically, the channel cilia of 3/3 che-12(mn389) animals examined lacked distal ciliary structures normally pervaded by singlet microtubules (Figure 2, J and K); however, the middle ciliary segment and the transition zone were intact in these animals, as indicated by the presence of doublet microtubules (Figure 2L). Three of three dyf-11(mn392) animals examined by EM lacked all recognizable ciliary structures (Figure 2, H and I), except for the proximal transition zone (data not shown).
Because dyf-11, but not che-12, is also required for AWC-mediated odortaxis, we hypothesized that the elaborate wing-like ciliary extensions of the AWC neuron might display structural abnormalities in dyf-11 animals, but remain normal in che-12 mutants. Indeed, fluorescence imaging revealed that dyf-11(mn392), but not che-12(mn399) animals, lack the wing-like ciliary extensions of AWC neurons (Figure 2, D–F).
Our morphological studies, therefore, suggest that che-12 is important for the assembly of distal ciliary structures, and that dyf-11 is absolutely required for cilium biogenesis.
CHE-12 is a conserved HEAT-repeat protein:
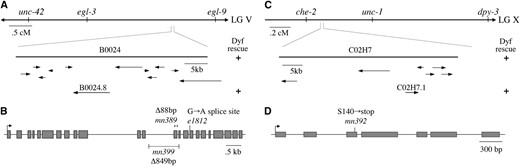
Previous genetic mapping experiments placed the che-12 gene in the interval between the unc-42 and daf-11 genes on chromosome V (Starichet al. 1995). We used SNPs between the che-12(mn389) and CB4856 strains to map che-12 to a smaller interval subtended by the C12D8 and AC3 cosmid clones (see materialsandmethods). Of the cosmids in this interval, only cosmid B0024 restored FITC dye uptake in amphid neurons when transformed into che-12(mn389) animals (three of three transgenic lines examined; Figure 3A), suggesting that this cosmid contained the che-12 gene. Indeed, a 9.2-kb subclone of cosmid B0024, containing only the B0024.8 gene, was sufficient for rescue (three of three lines examined). To confirm that B0024.8 was che-12, we sequenced the exons and exon/intron junctions of the gene and identified lesions in all three che-12 mutants described above (Figure 3B). The che-12(e1812) allele contained a G-to-A transition that disrupts the predicted splice-donor site of intron 16. The che-12(mn389) allele contained an 88-bp deletion, removing the last 47 bp of exon 13; and the che-12(mn399) allele harbored an 849-bp deletion, removing most of intron 12, exon 13, and the first 15 bp of exon 14, and is, therefore, likely to be the most defective che-12 allele of the three. Sequencing of another reported che-12 allele, e1813, showed that it contained the same lesion present in che-12(e1812) animals and was thus unlikely to be an independent isolate.
The genomic structures of che-12 and dyf-11. (A) che-12 was mapped to the indicated region of chromosome V. Rescue of the Dyf defect could be achieved by injection of the B0024 cosmid or the B0024.8 gene. (B) Genomic structure of B0024.8. The positions of the three alleles are shown. (C) dyf-11 was mapped to the indicated region of chromosome X. A single gene, C02H7.1, within cosmid C02H7, could rescue the Dyf defect. (D) Genomic structure of C02H7.1 and position of the mn392 allele.
To confirm the predicted che-12 gene structure (http://www.wormbase.org, release WS170), we isolated the che-12 cDNA (see materialsandmethods). Sequence analysis of this cDNA revealed that exon 10 is 291 bp longer than in the predicted transcript. By sequencing genomic DNA from the che-12 region of wild-type animals, we found that this misprediction arose because a C nucleotide at the beginning of intron 10 was not present in the annotated genomic sequence.
Analysis of the che-12 cDNA revealed that it encodes a protein of 1282 amino acids. A search of available protein databases using the BLAST program (Altschulet al. 1990) showed that the putative CHE-12 protein shares 34% similarity and 20% identity with a protein predicted to be encoded by human cDNA clone KIAA0423 (Ishikawaet al. 1997) and that has been detected by mass spectroscopy in human fetal brain (Hepneret al. 2005). Sequence analysis of the predicted CHE-12 protein suggests that it probably contains at least seven HEAT repeats, ∼39-aa-long elements that fold to form two antiparallel α-helices (Andrade and Bork 1995).
DYF-11 is similar to the mammalian microtubule-associated protein MIP-T3:
As with che-12, we used SNPs between the dyf-11(mn392) and CB4856 strains to map the dyf-11 gene to the interval between cosmid clones F39H12 and F02G3 on chromosome X (Figure 3C). This region contains 20 predicted genes, and sequencing of the predicted gene C02H7.1 from dyf-11(mn392) animals revealed that it contains a C-to-G mutation, altering codon 140, encoding serine, to an opal nonsense mutation (Figure 3D). To confirm that C02H7.1 was indeed dyf-11, we introduced the C02H7 cosmid, as well as a 4.1-kb DNA fragment containing only the C02H7.1 coding and regulatory sequences, into dyf-11(mn392) animals. Both transgenes rescued the dye-uptake defect (three transgenic lines examined for each transgene; Figure 3C). Together, these observations strongly suggest that C02H7.1 is dyf-11.
To decipher the dyf-11 gene structure, we isolated and sequenced a full-length cDNA for the gene (see materialsandmethods), confirming the predicted gene structure (http://www.wormbase.org, release WS170). Comparison of the predicted 535-aa-long DYF-11 protein to proteins in available databases showed that DYF-11 shares 43% similarity and 24% identity with the human protein MIP-T3 (microtubule-interacting protein that interacts with TRAF3, the tumor necrosis factor receptor-associated factor 3). MIP-T3 has been shown to interact with taxol-stabilized microtubules and with tubulin in vitro (Ling and Goeddel 2000), suggesting that DYF-11 may directly contact ciliary microtubules. Analysis of the DYF-11 protein sequence revealed two domains of note: a lysine-rich region between amino acids 122 and 214 and a predicted C-terminal coiled-coil region, a domain found in a number of IFT complex B proteins (Cole 2003), between amino acids 420 and 529.
che-12 is expressed in a subset of sensory neurons:
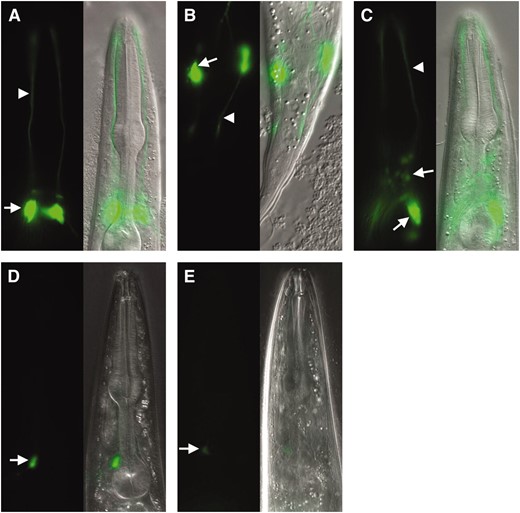
The phenotypic characterization of che-12 and dyf-11 suggested that both genes are likely to be expressed in sensory neurons. To test this idea, we determined the expression pattern of each gene. To examine che-12 expression, we generated a transgene bearing the 936-bp sequence present immediately upstream of the che-12 translation start site, fused to the gene encoding GFP, and introduced it into wild-type animals. This same 936-bp promoter fragment, which spans nearly the entire noncoding interval between the stop codon of the upstream gene, gcy-6, and the start codon of che-12, when driving expression of a che-12 cDNA∷GFP fusion (see below), was sufficient to rescue the dye-uptake defects of che-12(mn389) animals (three lines tested). Inspection of three independent che-12 pro∷GFP transgenic lines showed that che-12 is expressed in only the subset of amphid neurons that lack wing- or finger-like ciliary extensions (Figure 4A) and in the two phasmid neurons (Figure 4B). To confirm our identification of the che-12-expressing cells, we generated wild-type animals bearing two transgenes, a che-12 pro∷mCherry reporter and an osm-6 pro∷GFP reporter, which is expressed in most or all ciliated neurons (Colletet al. 1998). Overlap of expression was only seen in amphid and phasmid neurons (data not shown). These results are consistent with the structural and behavioral defects exhibited by che-12 mutants, and suggest that che-12 may only function in amphid neurons possessing simple cilia.
CHE-12 and DYF-11 are expressed in ciliated neurons in a DAF-19-dependent manner. (A) CHE-12 is expressed in a subset of amphid neurons. Neuronal cell bodies (arrow); dendritic processes (arrowhead). (B) CHE-12 is also present in phasmid neurons. (C) Expression of DYF-11 is seen in most ciliated neurons including those of the amphid and labial sensilla (arrows). (D) Expression of che-12 pro∷GFP is greatly reduced in daf-16; daf-19 animals. Compare with A. In D and E, image exposure was at least twice as long as in A and C. (E) The expression of dyf-11 pro∷GFP also depends on the transcription factor DAF-19. Compare with C.
dyf-11 is expressed in all ciliated sensory neurons:
To visualize the expression pattern of dyf-11, we used the 1868 bp immediately upstream of the dyf-11 translation start site to drive GFP expression in wild-type animals. GFP fluorescence was observed in most ciliated neurons, including neurons of the amphid, phasmid, and labial sensilla, as well as the neurons AQR, PQR, ADE, and PDE (Figure 4C). This result is consistent with a genomewide survey of ciliated neuron-expressed genes which demonstrated that C02H7.1 is expressed in the head and tail of C. elegans (Kunitomoet al. 2005). Our expression studies are also consistent with the dye-uptake and behavioral defects of dyf-11 mutants and suggest that dyf-11 plays an important role in the function of all cilia in C. elegans.
che-12 and dyf-11 expression is dependent on the transcription factor DAF-19:
In C. elegans, the expression of most genes encoding cilia-localized proteins requires DAF-19, an RFX transcription factor that recognizes a specific motif, the X box, in the promoter region of its target genes (Swobodaet al. 2000). To determine whether the transcription of che-12 and dyf-11 requires DAF-19, the che-12 pro∷GFP and dyf-11 pro∷GFP transgenes were introduced into daf-16(mu86); daf-19(m86) animals. daf-19(m86) mutants constitutively arrest in the dauer stage of development, a dormant, protective developmental stage normally entered under harsh environmental conditions (Riddle 1988). The daf-16(mu86) mutation suppresses the dauer arrest of daf-19(m86) mutants, allowing for the propagation of daf-19 mutant lines (Vowels and Thomas 1992). As shown in Figure 4, D and E, expression of both the che-12 pro∷GFP and dyf-11 pro∷GFP reporter transgenes was eliminated, or greatly reduced in daf-16(mu86); daf-19(m86) animals.
It has been previously noted that the C02H7.1 (dyf-11) promoter contains a consensus X-box sequence at position −194 with respect to the translation start site (Blacqueet al. 2005). The region upstream of the che-12 gene contains an X-box motif at position −1013 (Blacqueet al. 2005), however, this sequence is located within the upstream gene, gcy-6 and was not included in the promoter we used to drive GFP expression. While no consensus X-box sites were found in the 936-bp promoter fragment we used, we did find a sequence, ATCAGCTTGAAAAC, at position −767, that differs from the X-box consensus sequence, RTHNYYWTRRNRAC (Efimenkoet al. 2005), at only one position and that might be used to drive che-12 expression in the amphid and phasmid neurons.
These results strongly suggest that DAF-19, directly or indirectly, controls the expression of both che-12 and dyf-11.
CHE-12 and DYF-11 localize to cilia:
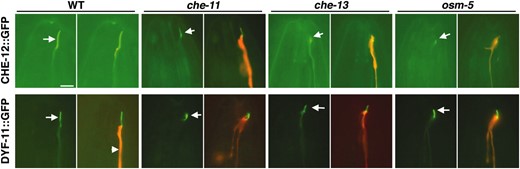
The mutant phenotypes and expression patterns of the che-12 and dyf-11 genes suggest that they may encode components of cilia. To test this hypothesis directly, we determined the localization of CHE-12∷GFP and DYF-11∷GFP fusion proteins. Specifically, we examined GFP localization in the ASER amphid neuron of animals expressing either che-12 cDNA∷GFP or dyf-11 cDNA∷GFP transgenes driven by the ASER-specific promoter of the gcy-5 gene (Yuet al. 1997). To visualize the ASER dendrite, mCherry was coexpressed from the same promoter, using an SL2 transsplicing acceptor sequence (see materialsandmethods). CHE-12∷GFP and DYF-11∷GFP localized throughout the ASER cilium (Figure 5) and were only faintly detectable in the ASER dendrite. Since both cDNA∷GFP fusion transgenes were able to rescue the Dyf phenotype of their respective mutants when expressed using their own promoters (data not shown), these results suggest that CHE-12 and DYF-11 proteins are normally targeted to neuronal sensory cilia, consistent with the ciliary defects observed in che-12 and dyf-11 mutants.
CHE-12 and DYF-11 localize to cilia. Left image of a pair, GFP alone. Right image, GFP and mCherry overlay. CHE-12 localizes to the cilium of ASER (arrows). The localization of CHE-12 is disrupted in che-13(1805) and osm-5(184) IFT-B mutants (note low intensity of GFP within cilium) and to a lesser extent in che-11(1810) IFT-A mutants. DYF-11 is normally localized within cilia of wild-type animals. This localization is not affected by the che-11, che-13, and osm-5 mutations. All transgenes are driven by the gcy-5 promoter, which is expressed specifically in ASER. Bar, 5 μm.
che-12 and dyf-11 act cell-autonomously within ciliated neurons:
che-12 and dyf-11 activities either could be required within a given ciliated neuron for its function or may influence the activities of nearby neurons nonautonomously. To distinguish between these two possibilities, we used the sra-6 promoter to express the che-12 and dyf-11 cDNAs in only two amphid neurons, ASH and ASI (Troemelet al. 1995), in che-12(mn389) and dyf-11(mn392) mutants, respectively. Only two neurons in each amphid were able to take up dye in these transgenic animals (Figure 6, B and F), suggesting that both che-12 and dyf-11 act cell autonomously.
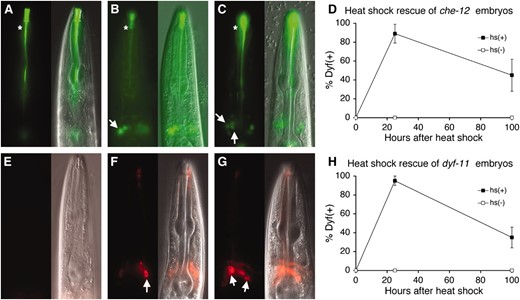
CHE-12 and DYF-11 act cell-autonomously and are required continuously for cilium morphology and function. (A) che-12(mn389) animals fail to take up FITC; asterisk indicates nonspecific staining. Fourth larval stage (L4) animals are depicted in all images. (B) Expression of CHE-12 in a che-12(mn389) mutant animal within only two amphid neurons, ASH and ASI, using the sra-6 promoter, rescues the Dyf defect only within these neurons (arrow). (C) A che-12(mn389) animal that is provided CHE-12 via heat shock as an adult is able to take up FITC in several amphid neurons (arrows). (D) che-12(mn389) embryos carrying extrachromosomal arrays containing the hsp-16.2 pro∷che-12 cDNA transgene were heat-shocked for 30 min at 34°. Dye filling was performed after 24 hr to determine initial rescue or after 100 hr. (E) dyf-11 animals cannot take up DiI. (F) Expression of dyf-11 in ASH and ASI using the sra-6 promoter enables only these two neurons to fill with dye (arrow). (G) Providing DYF-11 to adult animals via heat shock rescues their Dyf defect. Animals contain an hsp-16.2 pro∷dyf-11 cDNA transgene. (H) Same as D, except that dyf-11(mn392) mutants carrying an hsp-16.2 pro∷dyf-11 transgene were used.
che-12 and dyf-11 activities are required continuously for cilium function:
To determine the time of action of che-12 and dyf-11, we generated plasmids in which the cDNA of each gene was placed under the control of the hsp-16.2 heat-inducible promoter (Joneset al. 1986; Fireet al. 1990). Amphid neurons of che-12(mn389) or dyf-11(mn392) larvae or adults harboring these plasmids were unable to take up dye at 20° (Figure 6, A and E). However, dye uptake was restored to both mutant strains after a 30-min heat shock at 34° (Figure 6, C and G), suggesting that dyf-11 and che-12 can function at any time during the life of C. elegans to allow proper cilium function. Heat-shock expression was also sufficient to rescue the morphological defects of cilia, consistent with previous findings (Fujiwaraet al. 1999). Thus, 12 hr after heat shock ASER cilium length was 6.2 ± 0.5 μm (n = 10) in che-12(mn399) animals and 6.4 ± 0.6 μm (n = 10) in dyf-11 animals.
To determine whether expression of che-12 and dyf-11 is required continuously, we administered a heat shock during embryogenesis and examined dye uptake 24 hr and 100 hr later. The percentage of animals rescued for the dye-uptake defect declined with time (Figure 6, D and H), indicating that che-12 and dyf-11 are required both for development and for maintenance of a functional cilium.
CHE-12 may not be an IFT component, but requires IFT for its localization:
Some ciliary proteins undergo regular movements within cilia, a phenomenon termed IFT (Rosenbaum and Witman 2002; Scholey 2003). To determine whether this is the case for CHE-12 and DYF-11, we generated wild-type animals carrying extrachromosomal transgenic arrays containing low copy numbers of che-12 pro∷che-12∷GFP or dyf-11 pro∷dyf-11∷GFP reporter transgenes and examined GFP fluorescence dynamics in cilia using a spinning disk confocal microscope (see materialsandmethods). We were unable to see any movement of a rescuing CHE-12∷GFP fusion protein in amphid and phasmid neurons in 83 kymographs we generated. By comparison, movement of the IFT component CHE-13 was easily observed in 3/4 kymographs we examined (data not shown). These observations suggest that CHE-12 may not be a component of the IFT machinery and that it may not play a role in ciliary protein transport.
To further test the notion that CHE-12 is not a component of the IFT machinery, we examined the localization of IFT particle-associated proteins in che-12 mutants. The IFT particle is thought to consist of at least two subunits, A and B. We found that localization of both CHE-11∷GFP (associated with the IFT-A subunit) and CHE-13∷GFP (associated with the IFT-B subunit) was unaffected in che-12 mutants (Figure 7, B and E), consistent with the hypothesis that che-12 does not play a role in IFT.
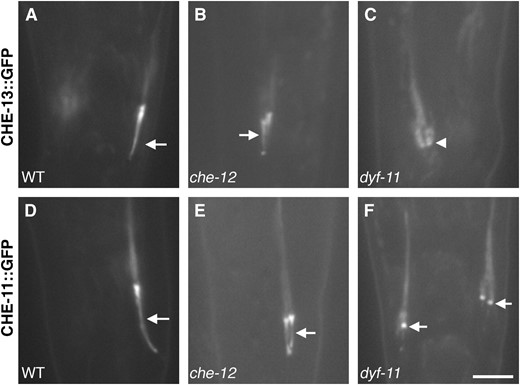
Localization of IFT-A and IFT-B components in che-12 and dyf-11 mutants. (A) Localization of CHE-13∷GFP in a phasmid cilium (arrow) of a wild-type animal. (B) In che-12(mn399) animals, CHE-13∷GFP localizes properly. (C) CHE-13∷GFP fails to localize to the cilia of dyf-11(mn392) animals; the base of the cilium is indicated by an arrowhead. (D) CHE-11∷GFP localization in a wild-type phasmid cilium. (E) In che-12(mn399) animals, CHE-11∷GFP localizes appropriately. (F) CHE-11∷GFP can occasionally localize to the base of the stunted cilia of dyf-11 animals. Bar, 4 μm.
Although CHE-12 is unlikely to be principally associated with IFT particles, its localization within the cilium may still depend on transport by IFT. To establish whether CHE-12 localization is IFT dependent, we examined the localization of ASER-expressed CHE-12∷GFP in IFT-defective mutants. The intensity of CHE-12∷GFP signal in the cilium was greatly reduced or absent in che-13 and osm-5 mutants (Figure 5), two components of the IFT-B complex (Haycraftet al. 2001, 2003; Qinet al. 2001). CHE-12∷GFP ciliary localization also appeared disrupted, but to a lesser extent, in the IFT-A complex mutant che-11 (Qinet al. 2001; Ouet al. 2005). These results suggest that CHE-12 at least partially requires IFT for its localization.
DYF-11 is associated with IFT particles:
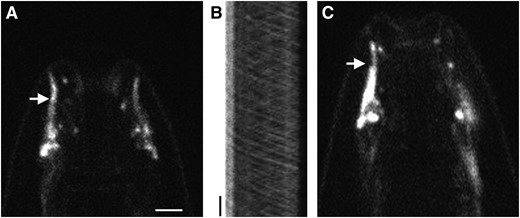
Unlike CHE-12∷GFP, a rescuing DYF-11∷GFP protein fusion displayed bidirectional movements within amphid cilia (Figure 8, A and B). Twenty-three of 62 kymographs we generated showed that DYF-11-containing particles moved in the anterograde direction along middle segments of amphid cilia with a rate of 0.79 ± 0.08 μm/sec and along distal segments with a rate of 1.19 ± 0.09 μm/sec. These velocities agree with those observed in the middle (0.7 μm/sec) and the distal segment (1.3 μm/sec) for other IFT proteins such as DYF-1, DYF-2, OSM-5, and OSM-6 (Snowet al. 2004; Ouet al. 2005; Efimenkoet al. 2006), suggesting that DYF-11 associates with IFT particles.
DYF-11 may associate with IFT particle B. (A) Localization of DYF-11∷GFP in the cilia (arrow) of a wild-type animal. (B) Kymograph of the cilia seen in A, showing anterograde and retrograde movement of DYF-11∷GFP. Position is displayed on the horizontal axis and time on the vertical axis. The transition zone of the cilium is on the left. Vertical bar, 5 sec. (C) DYF-11∷GFP can enter the distal segment of cilia in a bbs-8(nx77) mutant animal (arrow). Bar, A–C, 2.5 μm.
In bbs-8(nx77) mutants, the A and B IFT particle subunits dissociate and move independently, with only subunit B entering the distal segment of sensory cilia (Blacqueet al. 2004; Ouet al. 2005). We found that DYF-11∷GFP localizes throughout the cilia, including the distal segments, of bbs-8 mutants (Figure 8C). This observation suggests that DYF-11 may be a component of the IFT-B subunit, a hypothesis supported by the severely stunted cilia of dyf-11 mutants (Figure 2C), a feature of other IFT-B complex mutants (Perkinset al. 1986).
To place DYF-11 in the hierarchy of known IFT proteins, we studied its localization in three IFT-defective mutants. DYF-11 localized to cilia in che-11 (IFT-A), as well as in che-13 and osm-5 (IFT-B) animals (Figure 5). These observations indicate that the function of these proteins is dispensable for DYF-11 localization, and that DYF-11 probably associates with the IFT particle early in its assembly.
To test this idea further, we examined the localization of CHE-13∷GFP (Haycraftet al. 2003) and CHE-11∷GFP proteins (Qinet al. 2001) in dyf-11(mn392) animals. We found that CHE-13∷GFP (IFT-B) is not localized to phasmid cilia of dyf-11 animals (Figure 7C), supporting the idea that DYF-11 is part of the IFT-B subunit and that DYF-11 is likely to be recruited to the IFT particle before CHE-13. CHE-11∷GFP (IFT-A) fluorescence in the cilia of dyf-11 animals was also reduced (Figure 7F), consistent with an early role for DYF-11 in IFT particle assembly.
DISCUSSION
Cilia, present at the tips of sensory neuron dendrites and on the surfaces of most vertebrate cells (Wheatleyet al. 1996), have been the subject of intense study in recent years. Genomic and proteomic efforts to uncover a core group of cilia proteins has revealed the existence of several hundred proteins likely to comprise the cilium proteome, and at least some of these have been directly shown to reside within cilia (Avidor-Reisset al. 2004; Liet al. 2004; Blacqueet al. 2005; Efimenkoet al. 2005). Many of the ciliary proteins identified to date are components of the IFT machinery, composed of a multiprotein particle shuttled within the cilium by molecular motors of the kinesin and dynein families (Rosenbaum and Witman 2002; Scholey 2003). However, although the IFT mechanism may be complex, it is unlikely that all cilia proteins are components of this machinery. In addition to IFT components, structural proteins must exist that give cilia their shape, and signaling modules must be present that control information flow through the cilium. Here we have explored the roles of two genes, che-12 and dyf-11. Our observations suggest that whereas dyf-11 encodes a newly described IFT component, che-12 is likely to encode a protein that acts independently of the IFT machinery to control cilium function and morphology.
che-12 may be a non-IFT component of cilia:
In C. elegans, mutations affecting cilium morphology generally block the ability of some neurons of the amphid sensory organ to concentrate lipophilic dyes, such as DiI and FITC (Perkinset al. 1986). We show here that among Dyf mutants, che-12 mutants are unique in their ability to discriminate between the dyes FITC and DiI. Specifically, we show that all known che-12 mutants are defective in FITC uptake, but only partially blocked for the uptake of DiI. Although the mechanism of dye uptake in C. elegans is not known, it is thought to reflect a functional state of particular neurons, since not all neurons exposed to dye are able to concentrate it (Hedgecocket al. 1985). Thus, the defects of che-12 mutants suggest that mutations in the gene may affect only a subset of neuronal functions, suggesting that CHE-12 may not be a core component of the IFT machinery.
Several of our observations are consistent with this idea. First, CHE-12 protein localizes to cilia, but does so in a diffuse, non-particulate fashion, unlike many IFT components (Colletet al. 1998; Haycraftet al. 2003; Ouet al. 2005). Further, although we analyzed many cilia expressing a rescuing CHE-12∷GFP fusion protein, we were unable to detect any motion of this protein. We could, however, easily detect movements of CHE-13∷GFP (data not shown) and DYF-11∷GFP (Figure 8B).
Second, mutations in che-12 had no effect on the movement or localization of IFT components we examined, suggesting either that che-12 is not a component of the IFT complex or that it is peripherally associated with it and not required for its integrity.
Third, EM studies of all three che-12 alleles revealed that these animals lack part of the distal segment of at least some amphid sensory cilia (Figure 2 J–L; data not shown). This defect is somewhat reminiscent of defects in the cilia of animals carrying mutations in the IFT-A complex components che-11 and daf-10 (Perkinset al. 1986). However, CHE-12 is unlikely to be a component of the IFT-A particle as in che-11 and daf-10 mutants, large accumulations of IFT particles along the axoneme are visible (Qinet al. 2001), suggesting that the IFT-A complex plays a crucial role in retrograde transport within the cilium. This defect is not seen in che-12 mutants.
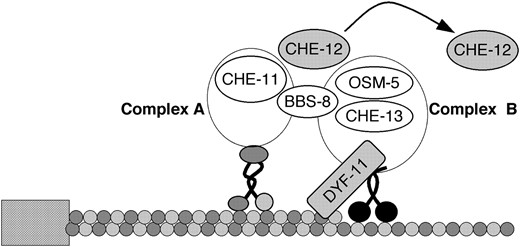
Although our evidence suggests that CHE-12 protein may be a non-IFT component of cilia, we show that its ciliary localization is dependent on a functioning IFT mechanism. We suggest a model, therefore, in which CHE-12 entry into cilia depends on IFT, but once inside cilia, the protein is released and free to provide IFT-independent functionality to these cellular compartments (Figure 9).
A hypothetical model showing the functions of CHE-12 and DYF-11 in sensory cilia. The IFT subcomplexes A and B, linked by the BBS-8 protein, are moved along the ciliary microtubules by the kinesin-2 and OSM-3 motors. DYF-11 might interact with microtubules as well as with the IFT-B protein complex, acting to promote association of CHE-13 and OSM-5 proteins with the complex. CHE-12 is transiently associated with the IFT particle to be transported into the cilium, where it is released and accumulates. Not all known IFT particle components are shown in this diagram.
che-12 may control the development and function of specific cilia:
Our studies of che-12 mutants reveal that these animals exhibit defects in only a subset of behaviors associated with amphid neurons. Specifically, behaviors associated with the olfactory neurons AWA and AWC seem generally unaffected in che-12 animals, consistent with the lack of che-12 expression in these neurons, whereas behaviors associated with neurons penetrating the amphid channel and responsive to soluble stimuli were defective. These observations suggest that che-12 may provide specialized functions to only a subset of amphid and phasmid neurons bearing simple cilia. It has already been noted that some cilia genes (osm-5, osm-6, che-2, che-13) are expressed in most or all ciliated neurons and are, therefore, required for the general formation of cilia (Efimenkoet al. 2005), while others, such as osm-3 (Tabishet al. 1995), dyf-3 (Murayamaet al. 2005), and dyf-2 (Efimenkoet al. 2006) are expressed in a subset of ciliated neurons and are hypothesized to lead to specialized ciliary structures (Mukhopadhyayet al. 2007). Genes restricted to only subsets of ciliated neurons seem to possess an asymmetric X-box sequence, the binding site of the ciliogenic transcription factor DAF-19, in their promoters (Swobodaet al. 2000). The che-12 promoter contains such an asymmetric X box (ATCAGC TT GAAAAC), further suggesting that CHE-12 might act to promote chemosensory cilia specialization.
We note that a previous EM study of che-12(e1812) animals reported that these animals have decreased levels of an undefined matrix in the amphid sheath cell, a glial cell that ensheaths the ciliated dendritic endings of the amphid neurons (Perkinset al. 1986), leading to the hypothesis that che-12 was likely to function in glia. However, both our studies of che-12 localization and our EM studies are inconsistent with these previous results.
How might CHE-12 function? Analysis of the CHE-12 protein sequence reveals that it contains seven or more HEAT repeats. These motifs, first described in the proteins huntigtin, elongation factor 3, PR65/A subunit of protein phosphatase 2A, and TOR1, are ∼39 residues long and are often repeated in tandem three or more times within a protein (Andrade and Bork 1995). Each HEAT motif folds as two antiparallel α-helices, and, as observed in the crystal structure of PR65/A (Groveset al. 1999), the helix pairs are arranged to form a solenoid with a hydrophobic inner core. In CHE-12, the first HEAT motif occurs at positions 99–135 while the last spans amino acids 1141–1177. Since most of the CHE-12 sequence is comprised of HEAT repeats, it is likely that the tertiary structure of CHE-12 resembles the solenoid formed by PR65/A. Such a structure could act as a scaffold and may be used to anchor ciliary proteins important both for FITC uptake and for building and stabilizing the distal segment of the cilium. The predicted human protein KIAA0423 shares the same HEAT-repeat topology as CHE-12, suggesting that it may also function within cilia in a manner similar to CHE-12.
DYF-11 protein is required early in IFT-B particle assembly:
While CHE-12 appears to be required for specialized cilia functions, our studies suggest that DYF-11 probably serves as a core IFT particle protein. Consistent with this idea, DYF-11 is expressed in most, if not all, ciliated neurons, and dyf-11 animals possess stunted cilia that resemble those of IFT-B class mutants. In addition, we could demonstrate that a DYF-11∷GFP rescuing fusion protein is able to undergo both anterograde and retrograde movements within cilia, at speeds consistent with those previously described for IFT.
The IFT complex can be dissociated into at least two subcomponents termed A and B. Previous studies have shown that in bbs-8 mutants, the IFT-A and IFT-B particles are separated, and IFT-A is transported along the medial segment via kinesin-2, while IFT-B is driven along the medial as well as the distal segment by OSM-3 (Ouet al. 2005). We found that DYF-11 localization to distal regions of cilia was unaffected in bbs-8 mutants, suggesting that DYF-11 is likely to be associated with the IFT-B subparticle.
Previous studies have suggested that the IFT particle is assembled in a series of steps. Analysis of IFT protein localization in different IFT mutant backgrounds has revealed some of this hierarchy of assembly. For example, OSM-5 is mislocalized in che-13 animals (Haycraftet al. 2003), indicating that CHE-13 acts prior to or together with OSM-5 in IFT complex B assembly. Furthermore, complex A proteins, such as CHE-11, are largely unaffected by removal of later acting IFT-B proteins. Interestingly, we showed that while DYF-11 localization was not disrupted in osm-5 (IFT-B), che-13 (IFT-B), or che-11 (IFT-A) mutants (Figure 5), localization of OSM-5 (data not shown), CHE-13, and CHE-11 was disrupted in dyf-11 animals (Figure 7, C and F). These results suggest that DYF-11 is likely to function early in IFT particle assembly and are consistent with a model in which DYF-11 is required for recruitment of OSM-5, CHE-13, and CHE-11 proteins to this particle (Figure 9).
DYF-11 could link signaling events to IFT in cilia:
Comparison of the DYF-11 protein sequence to existing protein databases revealed that this protein is conserved in evolution. Its human ortholog, MIP-T3, was identified in a yeast two-hybrid screen for factors that interact with TRAF3 (Ling and Goeddel 2000). MIP-T3 binds to taxol-stabilized microtubules and to tubulin in vitro and is able to recruit TRAF3 to microtubules in HeLa cell lines that overexpress both proteins (Ling and Goeddel 2000), raising the intriguing possibility that DYF-11 might be associated with ciliary microtubules (Figure 9). Although current models suggest that contact of the IFT complex with microtubules is mediated by the motors which move the complex, it is possible that a specific association of complex components with microtubules is required either for their initial assembly or to facilitate initiation or termination of IFT complex motion by its associated motors.
Studies of MIP-T3 also suggest that this protein interacts with the interleukin-13 receptor (IL-13Rα1) (Niuet al. 2003). Interestingly, the association of MIP-T3 with this receptor and with TRAF3 changes after signal transduction has occurred (Ling and Goeddel 2000; Niuet al. 2003). These observations suggest that DYF-11 may be in a position to control aspects of IFT in response to environmental cues received by cilia. The sequence similarity between DYF-11 and MIP-T3 also suggests that MIP-T3 may function in human primary cilia to control cytokine responses in target cells. Consistent with a potential regulatory role, MIP-T3 also interacts with Disrupted-in-Schizophrenia 1 (DISC1) and appears to be required for the recruitment of DISC1, a centrosome associated protein, to microtubules (Morriset al. 2003).
Primary cilia are found on most human cells, and their roles in many aspects of signal transduction are revealed by the pleiotropic defects observed in Bardet–Biedl syndrome (BBS) patients (Ansleyet al. 2003). Here we have described two conserved cilia proteins: CHE-12, which might act as a structural scaffold-promoting specialized structure and/or function of some cilia, and DYF-11, which acts as an IFT-B particle component. Further studies of these and other cilia components should help to reveal the mechanics of signal transduction and processing performed by this organelle.
Footnotes
Communicating editor: K. J. Kemphues
Acknowledgement
We thank Max Heiman for the unc-42 che-12 egl-9 strain and for first pointing out that che-12 mutants show differential dye uptake, Graeme Couture for help in cosmid preparation, Maya Goldmit for the SL2 operon construct, Alison North of the Rockefeller University Bio-Imaging Center for help with imaging, and members of the Shaham laboratory for helpful comments on the project and manuscript. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health. S.S. is a Klingenstein fellow in the neurosciences and a Monique Weill-Caulier scholar.
References
Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman,
Andrade, M. A., and P. Bork,
Ansley, S. J., J. L. Badano, O. E. Blacque, J. Hill, B. E. Hoskins et al.,
Avidor-Reiss, T., A. M. Maer, E. Koundakjian, A. Polyanovsky, T. Keil et al.,
Bargmann, C. I., and H. R. Horvitz,
Bargmann, C. I., E. Hartwieg and H. R. Horvitz,
Bell, L. R., S. Stone, J. Yochem, J. E. Shaw and R. K. Herman,
Blacque, O. E., M. J. Reardon, C. Li, J. McCarthy, M. R. Mahjoub et al.,
Blacque, O. E., E. A. Perens, K. A. Boroevich, P. N. Inglis, C. Li et al.,
Brenner, S.,
Cole, D. G.,
Collet, J., C. A. Spike, E. A. Lundquist, J. E. Shaw and R. K. Herman,
Culotti, J. G., and R. L. Russell,
Efimenko, E., K. Bubb, H. Y. Mak, T. Holzman, M. R. Leroux et al.,
Efimenko, E., O. E. Blacque, G. Ou, C. J. Haycraft, B. K. Yoder et al.,
Fire, A., S. W. Harrison and D. Dixon,
Fujiwara, M., T. Ishihara and I. Katsura,
Groves, M. R., N. Hanlon, P. Turowski, B. A. Hemmings and D. Barford,
Haycraft, C. J., P. Swoboda, P. D. Taulman, J. H. Thomas and B. K. Yoder,
Haycraft, C. J., J. C. Schafer, Q. Zhang, P. D. Taulman and B. K. Yoder,
Hedgecock, E. M., J. G. Culotti, J. N. Thomson and L. A. Perkins,
Hepner, F., J. K. Myung, N. Ulfig, A. Pollak and G. Lubec,
Hodgkin, J., and T. Doniach,
Huang, T., S. Kuersten, A. M. Deshpande, J. Spieth, M. MacMorris et al.,
Ishikawa, K., T. Nagase, D. Nakajima, N. Seki, M. Ohira et al.,
Jones, D., R. H. Russnak, R. J. Kay and E. P. Candido,
Kaplan, J. M., and H. R. Horvitz,
Kunitomo, H., H. Uesugi, Y. Kohara and Y. Iino,
Li, J. B., J. M. Gerdes, C. J. Haycraft, Y. Fan, T. M. Teslovich et al.,
Ling, L., and D. V. Goeddel,
Lundquist, E. A., P. W. Reddien, E. Hartwieg, H. R. Horvitz and C. I. Bargmann,
Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros,
Miller, D. M., III, N. S. Desai, D. C. Hardin, D. W. Piston, G. H. Patterson et al.,
Morris, J. A., G. Kandpal, L. Ma and C. P. Austin,
Mukhopadhyay, S., Y. Lu, H. Qin, A. Lanjuin, S. Shaham et al.,
Murayama, T., Y. Toh, Y. Ohshima and M. Koga,
Niu, Y., T. Murata, K. Watanabe, K. Kawakami, A. Yoshimura et al.,
Orozco, J. T., K. P. Wedaman, D. Signor, H. Brown, L. Rose et al.,
Ou, G., O. E. Blacque, J. J. Snow, M. R. Leroux and J. M. Scholey,
Pazour, G. J., and G. B. Witman,
Perens, E. A., and S. Shaham,
Perkins, L. A., E. M. Hedgecock, J. N. Thomson and J. G. Culotti,
Qin, H., J. L. Rosenbaum and M. M. Barr,
Riddle, D. L.,
Rosenbaum, J. L., and G. B. Witman,
Sarafi-Reinach, T. R., T. Melkman, O. Hobert and P. Sengupta,
Scholey, J. M.,
Scholey, J. M., and K. V. Anderson,
Schumacher, B., C. Schertel, N. Wittenburg, S. Tuck, S. Mitani et al.,
Signor, D., K. P. Wedaman, J. T. Orozco, N. D. Dwyer, C. I. Bargmann et al.,
Singla, V., and J. F. Reiter,
Snow, J. J., G. Ou, A. L. Gunnarson, M. R. Walker, H. M. Zhou et al.,
Starich, T. A., R. K. Herman, C. K. Kari, W. H. Yeh, W. S. Schackwitz et al.,
Swoboda, P., H. T. Adler and J. H. Thomas,
Tabish, M., Z. K. Siddiqui, K. Nishikawa and S. S. Siddiqui,
Troemel, E. R., J. H. Chou, N. D. Dwyer, H. A. Colbert and C. I. Bargmann,
Troemel, E. R., A. Sagasti and C. I. Bargmann,
Vowels, J. J., and J. H. Thomas,
Ward, S.,
Ward, S., N. Thomson, J. G. White and S. Brenner,
Wheatley, D. N., A. M. Wang and G. E. Strugnell,
White, J. G., E. Southgate, J. N. Thomson and S. Brenner,
Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk,
Yu, S., L. Avery, E. Baude and D. L. Garbers,