-
PDF
- Split View
-
Views
-
Cite
Cite
Amber M Hohl, Morgan Thompson, Alexey A Soshnev, Jianhong Wu, James Morris, Tao-Shih Hsieh, C-ting Wu, Pamela K Geyer, Restoration of Topoisomerase 2 Function by Complementation of Defective Monomers in Drosophila, Genetics, Volume 192, Issue 3, 1 November 2012, Pages 843–856, https://doi.org/10.1534/genetics.112.144006
Close - Share Icon Share
Abstract
Type II topoisomerases are essential ATP-dependent homodimeric enzymes required for transcription, replication, and chromosome segregation. These proteins alter DNA topology by generating transient enzyme-linked double-strand breaks for passage of one DNA strand through another. The central role of type II topoisomerases in DNA metabolism has made these enzymes targets for anticancer drugs. Here, we describe a genetic screen that generated novel alleles of DrosophilaTopoisomerase 2 (Top2). Fifteen alleles were obtained, resulting from nonsense and missense mutations. Among these, 14 demonstrated recessive lethality, with one displaying temperature-sensitive lethality. Several newly generated missense alleles carry amino acid substitutions in conserved residues within the ATPase, Topoisomerase/Primase, and Winged helix domains, including four that encode proteins with alterations in residues associated with resistance to cancer chemotherapeutics. Animals lacking zygotic Top2 function can survive to pupation and display reduced cell division and altered polytene chromosome structure. Inter se crosses between six strains carrying Top2 missense alleles generated morphologically normal trans-heterozygous adults, which showed delayed development and were female sterile. Complementation occurred between alleles encoding Top2 proteins with amino acid substitutions in the same functional domain and between alleles encoding proteins with substitutions in different functional domains. Two complementing alleles encode proteins with amino acid substitutions associated with drug resistance. These observations suggest that dimerization of mutant Top2 monomers can restore enzymatic function. Our studies establish the first series of Top2 alleles in a multicellular organism. Future analyses of these alleles will enhance our knowledge about the contributions made by type II topoisomerases to development.
TOPOLOGICAL changes in chromosome structure are resolved through the action of a wide variety of enzymes known as topoisomerases. Among these, type II topoisomerases are conserved enzymes that alter DNA structure by introducing a transient double-strand break (DSB) in one DNA strand and passing a second DNA strand through the cleaved strand (Wang 2002; Nitiss 2009a). This catalytic cycle involves the covalent attachment of topoisomerase II to DNA, a reversible reaction that ends by ligation of the DSB. Formation of an enzyme–DNA intermediate protects the DNA ends and prevents activation of a DNA damage checkpoint.
Type II topoisomerases are homodimeric proteins (Schoeffler and Berger 2008; Collins et al. 2009; Nitiss 2009a). Each monomer is composed of distinct domains that cooperate to alter DNA topology (Figure 1). The amino-terminal ATPase domain is responsible for ATP binding and hydrolysis, which promotes dimer formation and regulates DNA opening and closing. Flanking the ATPase domain is the Transducer domain (TDD), which signals ATP binding to the catalytic core. Following the TDD is the catalytic core, composed of two domains required for DNA breakage and religation. Of these, the Topoisomerase/Primase (TOPRIM) domain contains a triad of acidic amino acids that are required for the DNA cleavage reaction and the Winged helix domain (WHD) contains the active-site tyrosine, which forms the covalent linkage with DNA. The Tower domain (TD) and Coiled-coiled domain (CCD) follow the catalytic core and together regulate the passage of one DNA strand. Finally, the carboxyl-terminal domain (CTD) is dispensable for catalytic activity in vitro, but regulates nuclear accumulation and interactions with partner proteins in vivo (Collins and Hsieh 2009). Among these domains, the CTD is the least conserved among eukaryotes, differing both in length and in sequence (Austin et al. 1993). Structural domains within dimeric type II topoisomerases are formed by contributions of both monomers (Liu and Wang 1999; Classen et al. 2003), which facilitates coupling of ATP hydrolysis to conformational changes involved in altering DNA structure.
Structure of the Top2 locus. (A) Top2 is located on chromosome 2L between the uncharacterized upstream CG10026 gene and the essential downstream RanGap gene. Shown are the structures of three Top2 deficiency chromosomes used in these studies. Top2Df9043 is a 14.8-kb deletion allele (dashed line) that removes Top2, RanGap, Hs2st (the gene within the RanGap intron), and CG10237. Top2Df17 is an ∼3.6-kb deletion allele (dashed line) that removes only Top2 sequences and retains ∼600 bp of the starting P element (Top2EP). Top2Df35 is a deletion allele that removes Top2 sequences, but has unknown limits (dotted line). Promoters are indicated by bent arrows and exons are represented by shaded rectangles. (B) P[Top2-w+] is a P transposon that carries white+ (not shown), a 7.1-kb genomic fragment encompassing the entire Top2 gene, and the 5′ region of CG10026 and the 3′ largely untranslated region of RanGap. The coding region of the Top2 gene is annotated to indicate locations of the ATPase domain, the Transducer domain (TDD), the Topoisomerase/Primase (TOPRIM) domain, the Winged helix domain (WHD), the Tower domain (TD), the Coiled-coiled domain (CCD), and the carboxyl-terminal domain (CTD).
In light of the function of type II topoisomerases, it is not surprising that these enzymes are structurally conserved and encoded by essential genes (Nitiss 2009a). The yeast and Drosophila genomes each contain a single gene, called Topoisomerase 2 (Top2). Mammalian genomes contain two Top2 genes, Top2A and Top2B, which encode the differentially expressed paralogs Top2α and Top2β, respectively (Austin and Marsh 1998). Top2α is found in proliferating cells, while Top2β is found ubiquitously, with elevated levels in terminally differentiated cells (Capranico et al. 1992; Watanabe et al. 1994). Interestingly, yeast Top2 mutants are rescued by expression of the Drosophila or human Top2 protein (Wyckoff and Hsieh 1988; Jensen et al. 1996), illustrating the strong functional conservation among eukaryotic type II topoisomerases.
Eukaryotic type II topoisomerases resolve entwined DNA strands and relax supercoiled structures that arise from the action of DNA polymerases. Genetic knockdown and chemical inhibitor studies have revealed that loss of Top2 causes chromosome missegregation and DNA damage during mitosis due to a failure to resolve sister chromatids and centromeres (Chang et al. 2003; Baxter and Diffley 2008; Coelho et al. 2008; Gonzalez et al. 2011). Some of these defects may result from altered chromosome architecture (Uemura et al. 1987; Buchenau et al. 1993; Chang et al. 2003; Coelho et al. 2008; Stanvitch and Moore 2008), as Top2α is a major structural component of mitotic chromosomes (Earnshaw et al. 1985; Adachi et al. 1991; Maeshima and Laemmli 2003). A role in global chromosome architecture is further suggested by studies in Drosophila, where Top2 may be involved in somatic homolog pairing (Williams et al. 2007) and insulator function (Ramos et al. 2011). The essential mitotic requirement for type II topoisomerases has made these enzymes important targets for chemotherapy against a number of cancers (Nitiss 2009b; Chikamori et al. 2010).
Our interest in Drosophila Top2 began with an earlier analysis demonstrating that compromising the function of this protein perturbs homolog pairing in cell culture (Williams et al. 2007). Here, we describe a genetic screen to generate a series of ethyl methanesulfonate (EMS)-induced Top2 alleles, as we reasoned that hypomorphic and null mutations would provide a useful resource that would complement the extant Top2 P-element insertion alleles (Bloomington Stock Center). In total, we identified 15 new alleles. Among these, 14 demonstrated recessive lethality, with one of these mutations displaying temperature sensitivity. Lethal alleles resulted from nonsense and missense substitutions in the protein-coding region, representing the first non-deletion, non-insertional Top2 alleles generated in Drosophila. Missense substitutions were in key functional domains of Top2 and included changes in residues associated with resistance to chemotherapeutic drugs (Wu et al. 2011). Inter se crosses between lethal missense alleles uncovered interallelic complementation, wherein trans-heterozygous Top2 mutant adults were generated. These adults were morphologically normal, although these flies showed delayed development and were female sterile. Interallelic complementation extended to crosses of strains carrying alleles encoding drug-resistant analogs of Top2. Taken together, these findings suggest that dimerization of some defective subunits can restore the in vivo activity of Top2. In brief, we have generated a new resource for investigating Top2 function.
Materials and Methods
Drosophila stocks and culture conditions
Flies were maintained at 25° at 70% humidity on standard Drosophila cornmeal, yeast, sugar, and agar medium with ρ-hydroxybenzoic acid methyl ester as a mold inhibitor. All crosses were performed at 25°, unless otherwise specified. Df(2L)Exel9043 (Bloomington Stock Center, BL 7913) is a 14.8-kb deletion on chromosome 2 that removes several genes including Top2, RanGap, Hs2st, and a portion of CG10237 (FlyBase, http://flybase.bio.indiana.edu/; Figure 1A). We refer to this deletion as Top2Df9043. Top2 mutant alleles were carried over the balancer chromosome CyO-Df(2R)B80, y+ balancer (BL 4542), referred to as CyO, y+. The Top2Df17 and Top2Df35 deficiencies were generated by excision of an existing P element (Top2EP). Top2Df17 contains a 3580-bp deletion within the Top2 coding region, with 590 bp remaining of the P element (Figure 1A). The breakpoints of Top2Df35 have not been mapped.
Mutagenic screen for the identification of Top2 mutant alleles
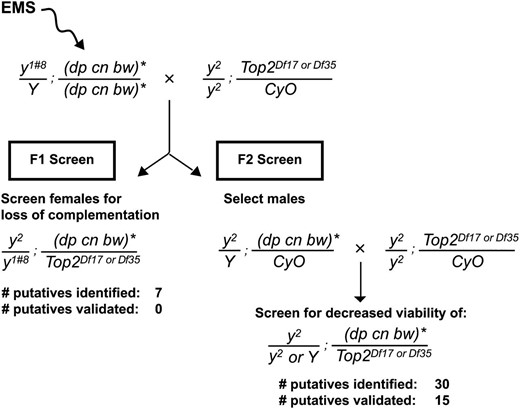
The strategy for isolating Top2 alleles is shown in Figure 2. Two- to four-day-old y1#8/Y; dp cn bw/dp cn bw males were desiccated for 12–14 hr and then fed 25 mM EMS in 10% sucrose (w/v). This mutagen was chosen as it largely produces nucleotide substitutions resulting from guanine alkylation (Bentley et al. 2000). After 24 hr of feeding, mutagenized males were transferred to bottles with standard food, allowed to recover for 10–12 hr, and mated for ∼24 hr with y2; Top2Df17 or Df35/CyO females. Mated females were transferred to fresh bottles daily for 3–4 days. F1 females of the genotype y2/y1#8; (dp cn bw)*/Top2Df17 or Df35 were scored for wing and body pigmentation. F1 males of the genotype y2/Y; (dp cn bw)*/CyO were singly mated in vials to y2/y2; Top2Df17 or Df35/CyO females. The resulting F2 progeny were screened, vial by vial, for decreased viability, as determined by an absence or reduced numbers of straight-winged flies. The y2/Y; (dp cn bw)*/CyO males from such vials were backcrossed to y2/y2; Top2Df17 or Df35/CyO females to confirm the decreased viability of straight-winged flies and to generate stocks carrying the putative Top2 mutations (Top2m). Stocks of putative Top2m mutations were established by subsequent balancing of the Top2m chromosome with the CyO, y+ chromosome, with 19 stocks established (Supporting Information, Table S2). Most crosses involved Top2m alleles balanced with the CyO, y+ chromosome; exceptional crosses involved Top2m alleles balanced with the CyO chromosome. These putative mutations were named based on the tester deficiency chromosome used in their identification, with those using the Top2Df17 tester named Top217-x and those using the Top2Df35 tester named Top235-x. The original tester deficiency chromosome for Top235-12 was unknown; this allele was arbitrarily assigned to the Top2Df35 group.
Strategy used in the EMS screen. Males of the genotype y1#8/Y; dp cn bw/dp cn bw were fed EMS and mated to y2/y2; TopDf17 or 35/CyO virgin females. In the F1 screen, y2/y1#8; (dp cn bw)*/TopDf17 or 35 females were screened for altered complementation between y alleles. In the F2 screen, F1 males carrying a mutagenized second chromosome in trans to the CyO balancer were crossed to virgin y2/y2; Top2Df17 or 35/CyO females. Vials were screened for absent or reduced numbers of straight-winged (Cy+) flies. * indicates the mutagenized chromosome. Of 3000 chromosomes screened, 30 putative Top2 alleles were identified, with 15 corresponding to bona fide alleles.
All putative mutations were rescreened by crossing y1w67c23; Top2m/CyO, y+ males or virgin females to flies carrying an independent Top2 deficiency chromosome (y1w67c23; Top2Df9043/CyO, y+). Crosses that produced no straight-winged flies were considered to contain new Top2 mutations. Next, we tested whether a wild-type Top2 gene rescued the recessive lethality associated with the Top2m alleles. To this end, we generated transgenic flies that carried an insertion of a genomic rescue transposon, P[Top2-w+], on the third chromosome. P[Top2-w+] carries a mini-white gene and a 7.1-kb HindIII to XbaI genomic fragment that includes the wild-type Top2 gene and part of the CG10026 gene (Figure 1B). As a control, we demonstrated that P[Top2-w+] rescued the recessive lethality associated with Top2Df17/Top2Df35 (Table S3), wherein Top2Df17/Top2Df35; P[Top2-w+]/+ individuals were produced at 86% of the Top2Df17 or Df35/CyO, y+ class.
Characterization of newly generated Top2 mutant alleles
Several studies were conducted to characterize the new Top2m alleles. First, we determined whether alleles displayed parent-of-origin effects by crossing y1w67c23; Top2m/CyO, y+ males or females to y1w67c23; Top2Df9043/CyO, y+ individuals of the opposite sex and scoring whether Top2m/Top2Df9043 adults survived. Second, we assessed whether Top2m alleles were temperature sensitive by crossing y1w67c23; Top2m/CyO, y+ females to y1w67c23/Y; Top2Df9043 /CyO, y+ males at room temperature for 2 days, after which time progeny were placed at 18°. Progeny emerging from these crosses were scored daily to determine whether growth at 18° generated Top2m/Top2Df9043 adults. Third, we assessed complementation between the Top2m alleles, performing inter se crosses between y1w67c23; Top2m/CyO, y+ or y2; Top2m/CyO mutants at 25°, with initial experiments scoring progeny every 2–3 days (data not shown). Complementation was observed between several different pairs of alleles, as Top2m1/Top2m2 adults were generated, which we call Top2-complementing adults. Crosses producing Top2-complementing adults were retested, with progeny screened daily. Fourth, we tested whether any of the Top2 mutations represented hypomorphic alleles. In these studies, we assessed partial Top2 function by examining viability of Top2m/Top2Df9043 larvae. Flies from each lethal Top2 mutant strain (y1w67c23/y1w67c23; Top2m/CyO, y+) were crossed to y1w67c23/Y; Top2Df9043/CyO, y+ flies and the y1w67c23; Top2m/Top2Df9043 second- and third-instar larvae were collected, using the absence of pigmentation in the mouth hooks and denticle belts to identify the correct genotype. For each cross, at least 100 larvae were selected and placed in vials, with no more than 25 larvae per vial, and allowed to develop at 25° under uncrowded conditions. The total numbers of individuals advancing to later stages were scored, with the percentage of survival at each stage determined by dividing the number of individuals obtained by the total number of larvae placed in each vial, multiplied by 100. Top2m alleles were considered hypomorphic if Top2m/Top2Df9043 larvae showed greater survival than did larvae from control crosses performed using confirmed null alleles. Fifth, the fertility of Top2 complementing males and females was assessed. In these studies, 8–10 y1 w67c23; Top2m1/Top2m2 males or females were crossed to 10–15 y1 w67c23; Sco/CyO flies of the opposite sex. Crosses were transferred every 2–3 days. Top2-complementing males were considered fertile if adults were produced from the cross. Top2-complementing females were considered fertile if they produced eggs that hatched. Crosses of flies from the parental line (y1#8; dp cn bw) were studied in parallel, as a control (data not shown).
Molecular characterization of Top2 alleles
Genomic DNA was isolated from heterozygous y1w67c23; Top2m/Top2Df9043 larvae and PCR amplified using primers covering nine overlapping regions encompassing the Top2 protein-coding region. PCR products were purified using the QIAquick PCR purification kit (QIAGEN, Valencia, CA; no. 28104) and sequenced at the University of Iowa DNA Core Facility. Primers are listed in Table S1. Western analysis was used to determine the level of Top2 protein generated in each strain. For these studies, proteins were extracted from a collection of brain, imaginal disc, and salivary gland tissues dissected from 10 Top2m/Top2Df9043 larvae. Two primary antibodies were used to detect Top2: rabbit anti-Top2 (T. Hsieh) at 1:10,000 (Figure 3) and rabbit anti-Top2 (D. Arndt-Jovin) at 1:4000 (data not shown). HRP-conjugated goat anti-rabbit IgG secondaries [Bio-Rad, Hercules, CA; no. 172-1019] were used at 1:20,000.
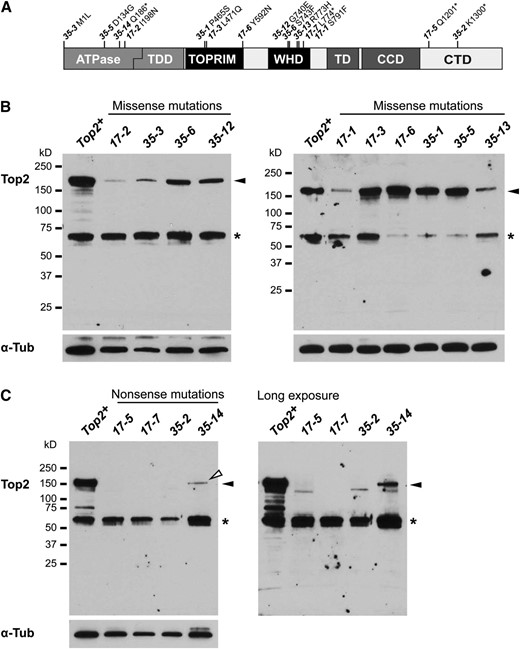
Analyses of protein production in Top2 mutations. (A) The locations of the nonsense and missense mutations in the lethal Top2 alleles are shown relative to the Top2 protein domain structure. Abbreviations are described in Figure 1. (B and C) Top2 protein levels were determined by Western analysis of proteins extracted from Top2m/Top2Df9043 third-instar larvae carrying missense (B) and nonsense (C) alleles. Westerns were probed with antibodies generated in the Hsieh laboratory. Antibodies against α-tubulin served as a loading control. One larval equivalent was loaded per lane. Solid arrowheads indicate the position of the full-length Top2. The open arrowhead marks the position of a full-length protein obtained from Top235-14/Top2Df9043 larvae, even though the Top235-14 nonsense allele is predicted to encode a protein of ∼20 kDa. The absence of a full-length band in protein extracts obtained from lines carrying other nonsense alleles suggests that this band represents context-specific readthrough of the nonsense codon. Asterisks mark the position of a cross-reacting band that was routinely observed.
Polytene chromosome and immunohistochemical analyses
Salivary gland polytene chromosomes were studied to define the impact of loss of Top2 on chromosome structure. Females of the genotype y1w67c23/y1w67c23; Top217-5 or 35-15/CyO, y+ were crossed to y1w67c23/Y; Top2Df9043/CyO, y+ males at room temperature for 2 days and then placed at 18°, according to previously described methods (Johansen et al. 2009). Polytene chromosomes stained with antibodies against MSL1, MLE, and JIL-1 were prepared using the conventional squash protocol, while those stained with H4K16ac were prepared using the acid-free method. For each genotype, squashes were done on at least 10 salivary gland pairs, dissected from larvae obtained from at least two independent crosses. Over 100 nuclei were examined for each genotype.
The effects of Top2 loss on imaginal disc and ovary development were studied. Tissues were dissected into 1× phosphate-buffered saline (PBS) solution and fixed in 3% paraformaldehyde (Electron Microscopy Sciences; no. 15710) for 20 min. Each immunohistochemical analysis was completed at least twice, with each analysis including tissues obtained from at least 8–10 individuals of each genotype. After tissues were blocked in PBT + 5% normal goat serum (Vector Laboratories, Burlingame, CA; no. S-1000), they were incubated overnight with primary antibodies at 4°. Following washes, tissues were stained with secondary antibodies for 2–3 hr and with DAPI (1 μg/ml in PBT) for 5–10 min. Samples were mounted in Vectashield (Vector Laboratories; H1000), imaged with a Zeiss (Thornwood, NY) 710 confocal microscope, and processed using ImageJ.
Primary antibodies used in the immunohistochemical analyses include rabbit anti-Phospho-Histone H3(pS10) (Epitomics; no. 1173-1) at 1:500, rabbit anti-H4K16ac at 1:50 (Active Motif; no. 39929), chicken anti-JIL-1 at 1:100 (K. Johansen), mouse anti-Lamin Dm0 [Developmental Studies Hybridoma Bank (DSHB), University of Iowa; ADL84.12] at 1:500, rabbit anti-Lamin Dm0 (P. Fisher), rabbit anti-MSL1 at 1:100 (M. Kuroda), rabbit anti-MLE at 1:100 (M. Kuroda), rabbit anti-Vasa (Santa Cruz; no. sc-30210) at 1:500, and mouse anti-Spectrin (DSHB; no. 3A9) at 1:50. The secondary Alexa Fluor (AF) antibodies (Invitrogen; Molecular Probes, Eugene, OR) were used at a 1:500 dilution, including AF488 goat anti-rabbit (A11008), AF568 goat anti-rabbit (A11011), AF488 goat anti-mouse (A11001), AF568 goat anti-mouse (A11004), AF488 goat anti-chicken (A11039), and AF568 goat anti-chicken (A11041).
Results and Discussion
An EMS screen identified new Top2 alleles
Our screen was designed to recover Top2 alleles in the F1 and F2 generations (Figure 2). In the F1 generation, we sought putative Top2 alleles by screening for alteration of a phenotype that depends on somatic homolog pairing, while in the F2 generation, putative Top2 alleles were identified by their failure to complement a chromosome carrying a deletion of Top2. For this mutagenesis, we used EMS, which was fed to males that were isogenic for a second chromosome carrying the wild-type Top2 gene and the dp, cn, and bw markers. Following EMS treatment, males were crossed to females bearing one of two deletion alleles of Top2, Top2Df17 or Top2Df35, in trans to the CyO balancer chromosome carrying a wild-type Top2 gene and the dominant Curly wing marker.
In the F1 generation, we looked for loss or enhancement of pairing-dependent gene expression of the X-linked yellow (y) cuticle pigmentation gene. In particular, F1 Cy+ females were examined for changes in complementation between a paternally contributed y1#8 allele, which lacks the y promoter and is a complete null, and a maternally contributed y2 allele, which has the y wing and body enhancers blocked by a chromatin insulator (Geyer et al. 1990; Morris et al. 1999). In a Top2+ background, y2/y1#8; (dp cn bw)*/Top2Df17orDf35 females were expected to show dark, nearly wild-type pigmentation because chromosome pairing permits the wing and body enhancers of y1#8 to act in trans on the promoter of y2. If a Top2 mutation was recovered that affected pairing, then altered wing and body pigmentation would be expected. Importantly, females carrying y2 have darkly pigmented bristles regardless of pairing status, which enabled us to distinguish any exceptional y2/y1#8; (dp cn bw)*/Top2Df17 or Df35 females with reduced pigmentation from y1#8/y1#8; (dp cn bw)*/Top2Df17 or Df35 females arising from nondisjunction of the X chromosome.
From 2653 F1 females, we identified 12 straight-winged females that had altered cuticle pigmentation. At least 4 of these females showed an unambiguous, uniform light cuticle color, resembling females in which y pairing had been disrupted (Chen et al. 2002; Ou et al. 2009). Seven of these exceptional females were successfully mated and all produced males with the light cuticle pigmentation characteristic of the y1#8 allele, which was presumed to have been inherited from their mother. The recovery of such males is consistent with the parental exceptional females being y2/y1#8; (dp cn bw)*/Top2Df17 or Df35, with such females carrying a mutant Top2 allele that encoded a protein that was unable to promote chromosome pairing but provided adult viability. Alternatively, it may be that these exceptional females arose from X chromosome nondisjunction, wherein nondisjunction of y2 in conjunction with loss of y1#8 generated homozygous y2 tissues except in the germline, which remained heterozygous for y1#8 and y2. Unfortunately, all subsequent crosses with the progeny of these exceptional females failed to recapitulate the reduced pigmentation of the original females.
In the F2 generation, we looked for recessive lethal or semilethal alleles of Top2. Here, F1y2/Y; (dp cn bw)*/CyO males, each representing a single mutagenized second chromosome, were singly crossed in vials to y2/y2; Top2Df17 or Df35/CyO females, representing two different tester Top2 deficiency chromosomes. Next, each vial was scored for the absence or reduced numbers of straight-winged progeny, consistent with the presence of a de novo lethal or semilethal Top2 allele. Of 3000 crosses, 30 putative Top2m alleles were identified. From these putatives, we successfully established 19 y1w67c23; Top2m/CyO, y+ stocks (Table S2), while 11 putatives either were sterile or produced unhealthy stocks that were subsequently lost. Each of the 19 putative alleles was tested in trans to Top2Df9043, a Top2 deficiency that was generated independently of Top2Df17 and Top2Df35, and a total of 15 were confirmed (Table S2). Fourteen demonstrated complete lethality in trans to Top2Df9043, while the remaining mutation, Top235-4, showed 25% of the expected class when transmitted maternally and 87% when transmitted paternally. To ensure that the lethality of the newly generated mutations was due to mutation of Top2, we tested whether the P[Top2-w+] transgene that carried a wild-type Top2 gene rescued the viability of Top2m/Top2Df35 flies. Of note, Top235-4 was not tested in this study, due to the high viability observed upon paternal transmission. In all cases, viable Top2m/Top2Df35; P[Top2-w+]/+ adults were obtained (Table S3), confirming that lethality was due to mutations in Top2. In total, 15 lethal or semilethal Top2 alleles were generated.
Several parameters contribute to gene mutability, including the length of the transcription unit, the size of the protein, and different tolerances among proteins for altered amino acid sequence. Among these, a recent screen using Drosophila demonstrated that the best predictors of mutability are length and conservation of the protein (Cooper et al. 2010). Based on the number of recessive lethal mutations identified after screening ∼3000 chromosomes, these investigators predicted that EMS generates one lethal mutation for every 73 evolutionarily conserved amino acids. Based on this information, we estimated that our screen should have generated ∼13 new Top2 alleles, as Drosophila Top2 is a 167-kDa protein that displays ∼67% similarity to human Top2. Our recovery of 14 lethal mutations is in remarkable agreement with the predicted value, arguing that our identification of a large number of new Top2 alleles reflects the size and high conservation of functional domains throughout the protein.
Top2 missense alleles carry amino acid substitutions in critical functional domains
To define the molecular lesions associated with the Top2 mutations, genomic sequences encompassing the coding region were PCR amplified and sequenced. As a reference, we sequenced the coding region of the Top2 gene from the parental strain, identifying two base pair changes relative to the sequence of the Top2 gene curated at NCBI; one is a silent change (+G3040A, E565E) and one produces a conservative amino acid substitution (+C5628T, A1401V) in a less conserved region of Top2 (Crenshaw and Hsieh 1993). These data indicate that the sequence of the Top2 coding region is largely unchanged between strains. As an additional control, we sequenced the Top2 coding region from four putative Top2 mutations that had been identified in our original screen, but were found to complement Top2Df9043 in secondary analyses (Table S2). We predicted that these alleles would be unlikely to harbor changes in Top2. As expected, the coding region in all four was identical to that in the parental strain (data not shown). Sequence analyses of the Top2 coding region in the 15 lethal or semilethal Top2 mutations revealed that 3 alleles carried nonsense mutations, 1 allele carried a missense and a nonsense mutation, 1 allele carried 3 missense mutations, and 10 alleles carried single missense mutations (Table 1). In all cases, missense alleles encoded Top2 proteins with one amino acid substitution in a residue conserved with human Top2α, with 6 alleles that showed alterations in residues found to be invariant among all sequenced eukaryotic type II topoisomerases. The missense mutations in our alleles were clustered in three functional domains, including the ATPase, WHD, and TOPRIM domains (Figure 3A). Based on structural information provided from studies of human Top2α (Wei et al. 2005) and etoposide bound to Top2β (Wu et al. 2011), we predict that Top235-5 may alter the ATP-binding pocket; Top217-3 may alter DNA binding; and Top235-1, Top235-6, and Top235-12 might display altered binding to the Top2 inhibitor etoposide.
Properties of Top2 alleles
| Allele name . | Amino acid changea . | Affected domainb . | Protein accumulationc . | Interallelic complementationd . |
|---|---|---|---|---|
| 17-1 | S791F | WHD | +/− | Yes |
| 17-2 | I198N | ATPase | +/− | No |
| 17-3 | L471Q | TOPRIM | +++ | Yes |
| 17-5 | L318Q, Q1201e | NA | +/− | No |
| 17-6 | Y592N | TOPRIM | +++ | Yes |
| 17-7 | L774e | NA | — | No |
| 35-1 | R197C, C347S, P465S | ATPase, TDD, TOPRIM | +++ | Yes |
| 35-2 | K1300e | NA | +/− | No |
| 35-3 | M1L | NA | + | No |
| 35-4 | T712I | WHD | ND | ND |
| 35-5 | D134G | ATPase | +++ | Yes |
| 35-6 | S743F | WHD | ++ | No |
| 35-12 | G740E | WHD | ++ | No |
| 35-13 | R773H | WHD | ++ | Yes |
| 35-14 | Q186e | NA | — | No |
| Allele name . | Amino acid changea . | Affected domainb . | Protein accumulationc . | Interallelic complementationd . |
|---|---|---|---|---|
| 17-1 | S791F | WHD | +/− | Yes |
| 17-2 | I198N | ATPase | +/− | No |
| 17-3 | L471Q | TOPRIM | +++ | Yes |
| 17-5 | L318Q, Q1201e | NA | +/− | No |
| 17-6 | Y592N | TOPRIM | +++ | Yes |
| 17-7 | L774e | NA | — | No |
| 35-1 | R197C, C347S, P465S | ATPase, TDD, TOPRIM | +++ | Yes |
| 35-2 | K1300e | NA | +/− | No |
| 35-3 | M1L | NA | + | No |
| 35-4 | T712I | WHD | ND | ND |
| 35-5 | D134G | ATPase | +++ | Yes |
| 35-6 | S743F | WHD | ++ | No |
| 35-12 | G740E | WHD | ++ | No |
| 35-13 | R773H | WHD | ++ | Yes |
| 35-14 | Q186e | NA | — | No |
ND, not determined; NA, not applicable.
Boldface type: invariant between human Top2α and -β, mouse Top2α and -β, and yeast Top2.
TDD, Transducer domain; TOPRIM, Topoisomerase/Primase domain; WHD, Winged helix domain.
Protein accumulation as assayed by Western analysis (+++, protein levels equal to wild type; ++, intermediate levels; +, low levels; +/−, very low levels; —, no protein relative to wild-type).
Complementation with other Top2 alleles.
Denotes nonsense mutation.
| Allele name . | Amino acid changea . | Affected domainb . | Protein accumulationc . | Interallelic complementationd . |
|---|---|---|---|---|
| 17-1 | S791F | WHD | +/− | Yes |
| 17-2 | I198N | ATPase | +/− | No |
| 17-3 | L471Q | TOPRIM | +++ | Yes |
| 17-5 | L318Q, Q1201e | NA | +/− | No |
| 17-6 | Y592N | TOPRIM | +++ | Yes |
| 17-7 | L774e | NA | — | No |
| 35-1 | R197C, C347S, P465S | ATPase, TDD, TOPRIM | +++ | Yes |
| 35-2 | K1300e | NA | +/− | No |
| 35-3 | M1L | NA | + | No |
| 35-4 | T712I | WHD | ND | ND |
| 35-5 | D134G | ATPase | +++ | Yes |
| 35-6 | S743F | WHD | ++ | No |
| 35-12 | G740E | WHD | ++ | No |
| 35-13 | R773H | WHD | ++ | Yes |
| 35-14 | Q186e | NA | — | No |
| Allele name . | Amino acid changea . | Affected domainb . | Protein accumulationc . | Interallelic complementationd . |
|---|---|---|---|---|
| 17-1 | S791F | WHD | +/− | Yes |
| 17-2 | I198N | ATPase | +/− | No |
| 17-3 | L471Q | TOPRIM | +++ | Yes |
| 17-5 | L318Q, Q1201e | NA | +/− | No |
| 17-6 | Y592N | TOPRIM | +++ | Yes |
| 17-7 | L774e | NA | — | No |
| 35-1 | R197C, C347S, P465S | ATPase, TDD, TOPRIM | +++ | Yes |
| 35-2 | K1300e | NA | +/− | No |
| 35-3 | M1L | NA | + | No |
| 35-4 | T712I | WHD | ND | ND |
| 35-5 | D134G | ATPase | +++ | Yes |
| 35-6 | S743F | WHD | ++ | No |
| 35-12 | G740E | WHD | ++ | No |
| 35-13 | R773H | WHD | ++ | Yes |
| 35-14 | Q186e | NA | — | No |
ND, not determined; NA, not applicable.
Boldface type: invariant between human Top2α and -β, mouse Top2α and -β, and yeast Top2.
TDD, Transducer domain; TOPRIM, Topoisomerase/Primase domain; WHD, Winged helix domain.
Protein accumulation as assayed by Western analysis (+++, protein levels equal to wild type; ++, intermediate levels; +, low levels; +/−, very low levels; —, no protein relative to wild-type).
Complementation with other Top2 alleles.
Denotes nonsense mutation.
We determined the levels of Top2 protein produced in the 14 Top2 strains that carry lethal alleles. Our Western analyses used two polyclonal antibodies raised against the full-length Top2 protein, which was especially important for optimizing the detection of mutant proteins obtained from strains carrying nonsense mutations. Similar results were obtained using both antibodies (Figure 3B, Table 1, and data not shown). We found that Top2 was at low levels or absent in 7 strains, including all 4 strains carrying nonsense mutations and 3 strains carrying missense mutations, while the remaining 7 strains carrying missense mutations displayed intermediate to high protein levels. In general, Top2 proteins that carried changes in the TOPRIM domain accumulated high levels of protein, while those with amino acid substitutions in the ATPase domain or WHD produced varying levels of protein. Top235-3 produces low levels of protein even though the translation initiation ATG codon is mutated. This protein may arise from a low frequency of initiation at the nearby in-frame ATG codon located 12 codons downstream of the bona fide initiator, leading to a loss of the first few amino acids in Top2. A low level of full-length Top2 was also found in protein extracts obtained from Top235-14/Top2Df9043 larvae, a surprising finding because Top235-14 carries a nonsense mutation at codon 186 and is predicted to generate a 20-kDa protein. The observed full-length protein might result from context-specific readthrough of the 186 nonsense codon, a possibility supported by the absence of this band in extracts obtained from other lines carrying nonsense mutations. Further, recent findings suggest that stop codon readthrough in Drosophila is more prevalent than previously realized (Jungreis et al. 2011). We were surprised by the low level of protein produced by the Top235-2 allele that carries a nonsense mutation of codon 1300 because a similarly sized truncated Drosophila Top2 protein was functional in vitro and complemented mutations of the yeast Top2 gene (Crenshaw and Hsieh 1993). A possible explanation may lie in the observation that a nuclear localization signal is present in the last 60 amino acids (Crenshaw and Hsieh 1993), amino acids that are lost in the Top235-2 protein. The absence of a nuclear localization signal in Drosophila may prevent Top235-2 nuclear entry and lead to protein degradation. Taken together, our Western analyses show that some of the lethal Top2 alleles accumulate high levels of protein. Previous studies in Saccharomyces cerevisiae have shown that lethality resulting from Top2 depletion and lethality caused by catalytic inactivation of Top2 result from different mechanisms (Baxter and Diffley 2008). Here we demonstrate that our collection of Top2 mutations includes those that produce no protein and those that produce near wild-type levels of a functionally compromised protein. Thus, our mutations have the capacity to elucidate how Top2 depletion and inactivity cause different mechanisms of lethality.
Top2 missense alleles include temperature-sensitive and hypomorphic mutations
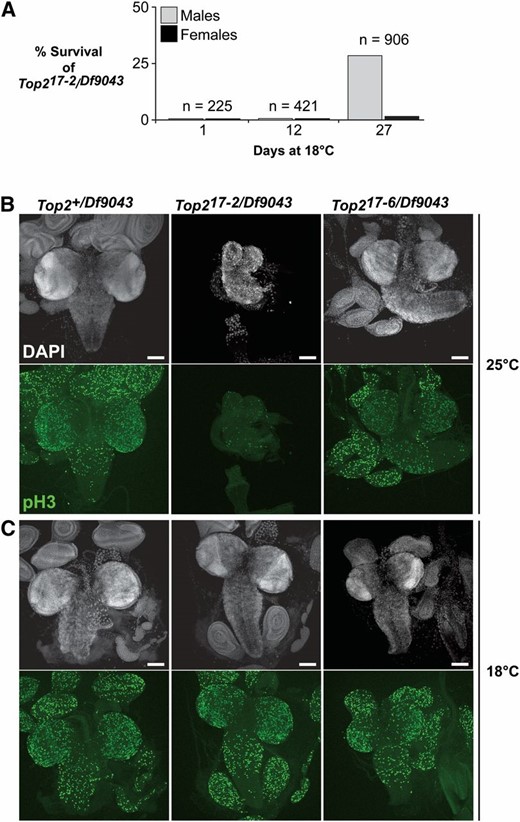
Temperature-sensitive alleles are a useful resource for in vivo functional studies. As missense mutations occasionally cause temperature sensitivity in protein function, we determined whether any of our Top2 mutations were temperature sensitive. We crossed y1w67c23/Y; Top2m/CyO, y+ females to y1w67c23; Top2Df9043/CyO, y+ males, collected eggs for 2 days at room temperature, and then transferred progeny to 18° for the remainder of development. These studies identified one genotype, Top217-2/Top2Df9043, that produced adults at the lower temperature. Based on the number of Top2m heterozygous siblings, we estimate that 29% of Top217-2/Top2Df9043 flies survived to adulthood (Table S2 and Figure 4A). Although Top217-2/Top2Df9043 females died shortly after emerging from their pupal cases, adult males survived for several days. To determine whether continued growth at 18° was essential for Top217-2/Top2Df9043 survival, y1w67c23/Y; Top217-2/CyO, y+ females were crossed to y1w67c23; Top2Df9043/CyO, y+ males at 18° and progeny were shifted to 25° at different developmental times. We found that a shift to 25° at 1 day (embryonic shift) or 12 days (pupal shift) resulted in a loss of adults (Figure 4A), suggesting that Top2 function is required both early and late in development, consistent with an essential function throughout development. Considering that Top217-2 encodes a full-length protein with a substitution in the highly conserved motif in the ATPase domain (I198N; Table 1), it is possible that the temperature sensitivity of Top217-2 results from defects associated with an inability of ATP binding or hydrolysis, properties needed to impart conformational changes needed to alter DNA topology. Alternatively, improved survival at the lower temperature may result from changes in Top217-2 accumulation, resulting from altered translation or degradation. Cold- and heat-sensitive Top2 mutants have been previously isolated in yeast (Thomas et al. 1991). These mutations resulted from missense mutations in conserved residues located outside of the ATPase domain and were clustered near the active site. These data suggest that missense mutations in multiple functional Top2 domains can generate temperature sensitivity.
Top217-2 is a temperature-sensitive allele. (A) Temperature-shift experiments revealed that one Top2 mutation, Top217-2, was temperature sensitive. Once identified, the developmental requirement for Top2 was determined by crossing Top217-2/CyO, y+ flies to Top2Df9043/CyO, y+ flies at 18° and shifting to 25° at 1, 12, or 27 days. The numbers of Top217-2/Top2Df9043 adults were counted, with percentage of survival estimated from the number of Top217-2 or Df9043/CyO, y+ siblings. Only Top217-2/Top2Df9043 individuals raised continuously at 18° reached adulthood, suggesting that Top2 is required throughout development. The Top217-2/Top2Df9043 females died shortly after eclosion. (B and C) Top2 is required for development of third-instar larval brains and imaginal discs. Tissues were isolated from Top2+/Top2Df9043, Top217-2/Top2Df9043, and Top217-6/Top2Df9043 mutants, carrying hypomorphic missense alleles. Larvae were raised at a nonpermissive (25°) or permissive (18°) temperature (B and C, respectively). Tissues were stained with the DNA stain DAPI (grayscale) and antibodies against Phospho-Histone H3(pS10) (green) to identify dividing cells. Images are shown as a maximum projection of a confocal Z-stack. Bar, 100 μm.
We were interested in defining whether our newly generated Top2 alleles represent complete or partial loss of function. To this end, we assessed the viability of Top2m/Top2Df9043 mutants, predicting that compared to complete loss of Top2 function, partial loss of function would permit increased survival. We crossed y1w67c23/y1w67c23; Top2m/CyO, y+ females to y1w67c23/Y; Top2Df9043/CyO, y+ males and followed development of y1w67c23; Top2m/Top2Df9043 larvae, identified by the absence of cuticle pigmentation. As a control, we tested two trans-heterozygous null genotypes (Top2Df17/Top2Df9043 and Top2Df35/Top2Df9043), anticipating that these genotypes would show extensive lethality prior to pupation, as previous studies have shown that Top2 alleles that produced undetectable levels of protein displayed high mortality in second-instar larvae, with <5% escaping into the third-instar larval stage (Ramos et al. 2011). Unexpectedly, we found that 49–66% of Top2Df17 or Df35/Top2Df9043 null larvae survived to pupation (Table 2), implying a lethal phase that is later than previously reported. Similar results were obtained for Top2m/Top2Df9043 mutant larvae carrying nonsense alleles, wherein 38–66% of mutant larvae survived to pupal stages (Table 2). Finally, we studied Top2m/Top2Df9043 larvae carrying Top2 missense alleles. We found that seven Top2m/Top2Df9043 genotypes demonstrated significantly enhanced survival relative to the known null genotypes, wherein 80–92% of mutant larvae became pupae (P < 0.04; Table 2). We note that one genotype, Top235-3/Top2Df9043 (P < 0.0002; Table 2) demonstrated significantly decreased survival relative to the null genotypes. Whether this difference reflects an unrelated influence of the genetic background or a special feature of Top235-3 is unclear. For example, Top235-3 may exert antimorphic effects, as it is predicted to carry a small amino-terminal deletion. Based on these data, we conclude that several newly identified missense mutations include hypomorphic Top2 alleles.
Analysis of survival of Top2m/Top2Df9043 larvae at 25°
| Allele namea . | Allele classification . | % pupationb . |
|---|---|---|
| Top2+ | Wild-type | 98b (100)c |
| Df 17 | Deletion | 49 (106) |
| Df 35 | Deletion | 66 (103) |
| 17-1 | Missense | 73 (100) |
| 17-2 | Missense | 72 (100) |
| 17-3 | Missense | 92 (120) |
| 17-5 | Missense, | 51 (142) |
| Nonsense | ||
| 17-6 | Missense | 80 (120) |
| 17-7 | Nonsense | 49 (135) |
| 35-1 | Missense | 82 (100) |
| 35-2 | Nonsense | 66 (110) |
| 35-3 | Missense | 26 (141) |
| 35-5 | Missense | 88 (100) |
| 35-6 | Missense | 83 (100) |
| 35-12 | Missense | 80 (100) |
| 35-13 | Missense | 82 (136) |
| 35-14 | Nonsense | 38 (104) |
| Allele namea . | Allele classification . | % pupationb . |
|---|---|---|
| Top2+ | Wild-type | 98b (100)c |
| Df 17 | Deletion | 49 (106) |
| Df 35 | Deletion | 66 (103) |
| 17-1 | Missense | 73 (100) |
| 17-2 | Missense | 72 (100) |
| 17-3 | Missense | 92 (120) |
| 17-5 | Missense, | 51 (142) |
| Nonsense | ||
| 17-6 | Missense | 80 (120) |
| 17-7 | Nonsense | 49 (135) |
| 35-1 | Missense | 82 (100) |
| 35-2 | Nonsense | 66 (110) |
| 35-3 | Missense | 26 (141) |
| 35-5 | Missense | 88 (100) |
| 35-6 | Missense | 83 (100) |
| 35-12 | Missense | 80 (100) |
| 35-13 | Missense | 82 (136) |
| 35-14 | Nonsense | 38 (104) |
Boldface type indicates hypomorphic alleles, strains that showed significant viability (P < 0.04) relative to both Top2Df17 and Top2Df35.
Maternally transmitted mutant Top2m, paternally transmitted mutant Top2Df9043.
Percentage of pupation was determined by dividing the total number of observed pupae by the total number of larvae placed in each vial, multiplied by 100.
Total number scored.
| Allele namea . | Allele classification . | % pupationb . |
|---|---|---|
| Top2+ | Wild-type | 98b (100)c |
| Df 17 | Deletion | 49 (106) |
| Df 35 | Deletion | 66 (103) |
| 17-1 | Missense | 73 (100) |
| 17-2 | Missense | 72 (100) |
| 17-3 | Missense | 92 (120) |
| 17-5 | Missense, | 51 (142) |
| Nonsense | ||
| 17-6 | Missense | 80 (120) |
| 17-7 | Nonsense | 49 (135) |
| 35-1 | Missense | 82 (100) |
| 35-2 | Nonsense | 66 (110) |
| 35-3 | Missense | 26 (141) |
| 35-5 | Missense | 88 (100) |
| 35-6 | Missense | 83 (100) |
| 35-12 | Missense | 80 (100) |
| 35-13 | Missense | 82 (136) |
| 35-14 | Nonsense | 38 (104) |
| Allele namea . | Allele classification . | % pupationb . |
|---|---|---|
| Top2+ | Wild-type | 98b (100)c |
| Df 17 | Deletion | 49 (106) |
| Df 35 | Deletion | 66 (103) |
| 17-1 | Missense | 73 (100) |
| 17-2 | Missense | 72 (100) |
| 17-3 | Missense | 92 (120) |
| 17-5 | Missense, | 51 (142) |
| Nonsense | ||
| 17-6 | Missense | 80 (120) |
| 17-7 | Nonsense | 49 (135) |
| 35-1 | Missense | 82 (100) |
| 35-2 | Nonsense | 66 (110) |
| 35-3 | Missense | 26 (141) |
| 35-5 | Missense | 88 (100) |
| 35-6 | Missense | 83 (100) |
| 35-12 | Missense | 80 (100) |
| 35-13 | Missense | 82 (136) |
| 35-14 | Nonsense | 38 (104) |
Boldface type indicates hypomorphic alleles, strains that showed significant viability (P < 0.04) relative to both Top2Df17 and Top2Df35.
Maternally transmitted mutant Top2m, paternally transmitted mutant Top2Df9043.
Percentage of pupation was determined by dividing the total number of observed pupae by the total number of larvae placed in each vial, multiplied by 100.
Total number scored.
Lethality during pupal stages of development suggested that Top2 mutants might carry defects in imaginal discs, which are ultimately responsible for generating adult tissues. In particular, we surmised that mutations in Top2 might interfere with the growth of imaginal discs, as previous studies in Drosophila cell culture have demonstrated that chemical inhibitors of Top2 decrease mitosis (Coelho et al. 2008). To test this postulate, we studied imaginal disc tissues isolated from multiple Top2 mutant strains, which revealed that imaginal discs and brains were larger in strains carrying hypomorphic alleles than in strains carrying null alleles. For example, Top217-6/Top2Df9043 larvae that produce high levels of mutant Top2 protein contain near wild-type–sized brains and small discs (Figures 3B and 4B), although these tissues display an altered cellular organization (data not shown). In contrast, Top217-2/Top2Df9043 larvae that produce very low levels of a mutant protein contain small brains and no imaginal discs when grown at the nonpermissive temperature, but larger brains and discs when grown at the permissive temperature (Figures 3B and 4B). To assess the level of mitosis in brains and imaginal discs obtained from Top217-6/Top2Df9043 and Top217-2/Top2Df9043 larvae, dissected tissues were stained with an antibody against phosphorylated serine 10 of histone H3, which is a highly selective marker for mitotic cells (Hendzel et al. 1997). These experiments revealed that the reduced size of brains and discs correlates with decreased staining of phosphorylated serine 10 of histone H3 (Figure 4, B and C), consistent with reduced levels of mitosis. Taken together, these data suggest that defects in imaginal disc growth may contribute to pupal lethality.
Loss of Top2 alters polytene chromosome structure
We studied the polytene chromosomes isolated from larval salivary glands to examine how loss of Top2 affects chromosome structure. These chromosomes contain ∼1000 aligned DNA strands, providing a powerful system for directly visualizing interphase chromosomes. Here we studied two null or nearly null Top2 genotypes, Top217-5/Top2Df9043 and Top235-14/Top2Df9043. Polytene chromosomes isolated from these female and male Top2 mutant larvae were fragile and displayed a diffuse DNA-banding pattern (Figure 5). This phenotype was particularly apparent for the male X chromosome, which displayed a “puffy,” decondensed structure, reminiscent of the structure associated with mutations in genes encoding chromatin-modifying proteins (Figure 5, A–C and A′–C′) (Deuring et al. 2000; Wang et al. 2001; Badenhorst et al. 2002; Spierer et al. 2005; Furuhashi et al. 2006; Bai et al. 2007; Grau et al. 2008). These observations suggest that loss of Top2 affects global chromosome structure, with the male X showing the greatest sensitivity within the genome.
Loss of Top2 affects chromosome structure. Salivary gland polytene chromosomes were prepared from male third-instar larvae representing wild-type [Top2+/Top2Df9043 (A, D, and G)] or mutant [Top217-5/Top2Df9043 (B, E, and H) and Top235-14/Top2Df9043 (C, F, and I)] Top2 genotypes. Chromosomes were stained with DAPI (A–C), with enlargement of the male X chromosome shown to demonstrate the diffuse DNA-banding pattern (A′–C′). Recruitment of the MSL complex was assessed using antibodies specific to MSL-1 (D–F) and H4K16ac (G–I).
The response of the male X chromosome to Top2 loss suggested a connection to the dosage compensation (DC) pathway. A possible connection to the DC pathway was further supported by the shared phenotype associated with hypomorphic mutations of JIL-1, which encodes a kinase component of the MSL complex that is responsible for interphase phosphorylation of histone H3 Ser10 in interphase cells (Deng et al. 2005). The dosage compensation pathway is required for increasing transcription from the single male X through recruitment of the Male Specific Lethal (MSL) complex, which leads to increased acetylation of lysine 16 of histone H4 (H4K16ac) (Gelbart and Kuroda 2009; Conrad and Akhtar 2011). To investigate whether the puffy X phenotype in Top2 mutant males resulted from altered dosage compensation, we examined X chromosome localization of components of the MSL complex in Top217-5/Top2Df9043 and Top235-14/Top2Df9043 male larvae. We found that loss of Top2 neither affects recruitment of MSL1, MLE, or JIL-1 nor alters enrichment of H4K16ac on the male X chromosome (Figure 5, D–F and G–I; data not shown).
Previous studies demonstrated that Imitation Switch (ISWI) regulates higher-order chromatin structure that includes global decondensation of salivary gland polytene chromosomes, with the male X chromosome displaying increased sensitivity relative to the autosomes (Deuring et al. 2000). These processes are linked to defects in nucleosome assembly due to reduced levels of histone H1 (Corona et al. 2007; Siriaco et al. 2009). Based on the similarity between the Top2 and Iswi mutant phenotypes, coupled with the previously recognized functional interaction between Top2 and H1 (Hsieh and Brutlag 1980), we reasoned that changes in polytene chromosome structure in Top2 mutants might result from changes in H1 levels. To this end, salivary gland proteins were isolated from Top2+/Top2Df9043, Top217-5/Top2Df9043, and Top235-14/Top2Df9043 larvae and assessed for H1 levels, using Western analyses. These studies demonstrated that H1 accumulates at near wild-type levels (data not shown), suggesting that loss of Top2 does not affect H1 production. It remains possible that loss of Top2 alters H1 deposition, a possibility that we were unable to test due to the inability of our H1 antibodies to work in immunohistochemical analyses.
Top2 mutations display interallelic complementation
Previous in vitro studies have demonstrated that Top2 subunits can undergo cooperative interactions to overcome defective ATP binding by one subunit (Lindsley and Wang 1993). Based on these findings, we predicted that missense alleles encoding Top2 proteins with mutations in distinct functional domains might display interallelic complementation. To this end, we conducted pairwise complementation tests between all Top2 alleles, assaying each cross for the production of viable adults. Initial crosses determined that 6 of the 14 lethal alleles supported interallelic complementation, generating Top2m1/Top2m2 adults (data not shown). We refer to these progeny as Top2-complementing adults. To determine the extent of complementation, we retested the 6 alleles, conducting crosses in both directions and scoring progeny daily (Table 3). Twenty-six crosses produced adults, revealing 13 heteroallelic genotypes capable of interallelic complementation. Based on the number of Top2m heterozygous siblings obtained in each complementation cross, we estimate that the viability of Top2m1/Top2m2 adults ranged from ∼13% to 98% (Table 3). Over half of all mutant genotypes displayed >50% viability of Top2m1/Top2m2 adults, indicating that interallelic complementation is robust.
Interallelic complementation between Top2 mutants y1w67c23/Y; Top2m/CyO, y+ × y1w67c23/ y1w67c23; Top2m/CyO, y+
| . | Allele transmitted by female . | |||||
|---|---|---|---|---|---|---|
| Allele transmitted by male . | 17-1 WHD . | 17-3 TOPRIM . | 17-6 TOPRIM . | 35-1a TOPRIM . | 35-5 ATPase . | 35-13 WHD . |
| 17-1 | 0b (165)c | 19 (275) | 13 (354) | 56 (612) | 95 (217) | 0 (220) |
| WHD | ||||||
| 17-3 | 22 (932) | 2 (163) | 64 (311) | 22 (719) | 93 (323) | 77 (183) |
| TOPRIM | ||||||
| 17-6 | 26 (214) | 52 (243) | 0 (248) | 58 (708) | 3 (147) | 76 (208) |
| TOPRIM | ||||||
| 35-1 | 46 (557) | 31 (705) | 71 (627) | 0 (98) | 78 (226) | 85 (360) |
| TOPRIM | ||||||
| 35-5 | 85 (220) | 88 (440) | 2 (777) | 86 (346) | 0 (103) | 64 (426) |
| ATPase | ||||||
| 35-13 | 0 (184) | 60 (350) | 55 (268) | 79 (655) | 98 (240) | 0 (275) |
| WHD | ||||||
| . | Allele transmitted by female . | |||||
|---|---|---|---|---|---|---|
| Allele transmitted by male . | 17-1 WHD . | 17-3 TOPRIM . | 17-6 TOPRIM . | 35-1a TOPRIM . | 35-5 ATPase . | 35-13 WHD . |
| 17-1 | 0b (165)c | 19 (275) | 13 (354) | 56 (612) | 95 (217) | 0 (220) |
| WHD | ||||||
| 17-3 | 22 (932) | 2 (163) | 64 (311) | 22 (719) | 93 (323) | 77 (183) |
| TOPRIM | ||||||
| 17-6 | 26 (214) | 52 (243) | 0 (248) | 58 (708) | 3 (147) | 76 (208) |
| TOPRIM | ||||||
| 35-1 | 46 (557) | 31 (705) | 71 (627) | 0 (98) | 78 (226) | 85 (360) |
| TOPRIM | ||||||
| 35-5 | 85 (220) | 88 (440) | 2 (777) | 86 (346) | 0 (103) | 64 (426) |
| ATPase | ||||||
| 35-13 | 0 (184) | 60 (350) | 55 (268) | 79 (655) | 98 (240) | 0 (275) |
| WHD | ||||||
Boldface type, 11–100% viability. Vials were scored daily.
Top235-1 has three amino acid changes; however, only one change is in an invariant amino acid (P465S in TOPRIM domain).
Percentage of viability is the number of Cy+ flies divided by half the number of Cy− flies multiplied by 100.
Total number of Cy− flies scored.
| . | Allele transmitted by female . | |||||
|---|---|---|---|---|---|---|
| Allele transmitted by male . | 17-1 WHD . | 17-3 TOPRIM . | 17-6 TOPRIM . | 35-1a TOPRIM . | 35-5 ATPase . | 35-13 WHD . |
| 17-1 | 0b (165)c | 19 (275) | 13 (354) | 56 (612) | 95 (217) | 0 (220) |
| WHD | ||||||
| 17-3 | 22 (932) | 2 (163) | 64 (311) | 22 (719) | 93 (323) | 77 (183) |
| TOPRIM | ||||||
| 17-6 | 26 (214) | 52 (243) | 0 (248) | 58 (708) | 3 (147) | 76 (208) |
| TOPRIM | ||||||
| 35-1 | 46 (557) | 31 (705) | 71 (627) | 0 (98) | 78 (226) | 85 (360) |
| TOPRIM | ||||||
| 35-5 | 85 (220) | 88 (440) | 2 (777) | 86 (346) | 0 (103) | 64 (426) |
| ATPase | ||||||
| 35-13 | 0 (184) | 60 (350) | 55 (268) | 79 (655) | 98 (240) | 0 (275) |
| WHD | ||||||
| . | Allele transmitted by female . | |||||
|---|---|---|---|---|---|---|
| Allele transmitted by male . | 17-1 WHD . | 17-3 TOPRIM . | 17-6 TOPRIM . | 35-1a TOPRIM . | 35-5 ATPase . | 35-13 WHD . |
| 17-1 | 0b (165)c | 19 (275) | 13 (354) | 56 (612) | 95 (217) | 0 (220) |
| WHD | ||||||
| 17-3 | 22 (932) | 2 (163) | 64 (311) | 22 (719) | 93 (323) | 77 (183) |
| TOPRIM | ||||||
| 17-6 | 26 (214) | 52 (243) | 0 (248) | 58 (708) | 3 (147) | 76 (208) |
| TOPRIM | ||||||
| 35-1 | 46 (557) | 31 (705) | 71 (627) | 0 (98) | 78 (226) | 85 (360) |
| TOPRIM | ||||||
| 35-5 | 85 (220) | 88 (440) | 2 (777) | 86 (346) | 0 (103) | 64 (426) |
| ATPase | ||||||
| 35-13 | 0 (184) | 60 (350) | 55 (268) | 79 (655) | 98 (240) | 0 (275) |
| WHD | ||||||
Boldface type, 11–100% viability. Vials were scored daily.
Top235-1 has three amino acid changes; however, only one change is in an invariant amino acid (P465S in TOPRIM domain).
Percentage of viability is the number of Cy+ flies divided by half the number of Cy− flies multiplied by 100.
Total number of Cy− flies scored.
Several conclusions were made from our inter se crosses. First, none of the nonsense alleles demonstrated interallelic complementation, suggesting that complementation requires full-length proteins. Second, alleles capable of interallelic complementation produced intermediate to high levels of Top2 protein (Table 1 and Figure 3). Even so, an absolute correlation between protein levels and complementation was not observed. For example, Top217-1 complemented several alleles, even though Top217-1 produces very low levels of protein, while Top235-6 failed to complement any allele, but produced intermediate levels of protein (Table 1 and Figure 3). Third, interallelic complementation occurred between alleles encoding proteins altered in different functional domains. For example, Top2 alleles encoding proteins with defects in the TOPRIM domain complemented alleles encoding proteins with defects in the ATPase domain or WHD (Table 3). Fourth, complementation was observed between alleles encoding proteins with defects in the same domain. Remarkably, complementation occurred between Top2 mutants carrying amino acid substitutions separated by only six residues in the TOPRIM domain (Top217-3/Top235-1). These studies provide the first demonstration that Top2 mutants can form a functional heterodimer in vivo and suggest that significant cooperation can occur between defective Top2 subunits to restore in vivo function.
Top2-complementing adults show limited developmental defects
We studied progeny from inter se crosses between Top2 missense alleles to understand the extent of restored Top2 function. In general, we found that Top2-complementing adults emerged at least 1 day later and died earlier than did their wild-type siblings. Nonetheless, these adults were morphologically normal, including having a body size that was similar to that of Top2+ flies. Unexpectedly, the F1 male-to-female ratio in 5 of 26 crosses showed significant deviation from the expected 1:1 ratio, wherein a single sex represented ≥70% of the offspring (Table S4). Higher numbers of male offspring were observed in all but one case (Top217-3/Top217-6). These findings are reminiscent of our observations that Top217-2/Top2Df9043 males produced by growth at 18° are healthier than Top217-2/Top2Df9043 females (Figure 4A). The reason for the different viability of Top2-complementing males and females is unclear.
We assessed the fertility associated with Top2-complementing adults. In initial studies, fertility was judged by an ability of Top2-complementing males or females to generate adult progeny when mated to y1 w67c23; Sco/CyO flies. This assay established that male fertility was largely not affected, with the one exception representing the heteroallelic genotype Top217-3/Top235-1 (Table 4). In contrast, all Top2-complementing females were sterile. To investigate the cause of female sterility, nonvirgin Top2-complementing females were placed in vials, which were examined for egg production. We found that all Top2m1/Top2m2 females produced low numbers of eggs. None of these eggs hatched, although their appearance was normal. As activation of a meiotic checkpoint is linked to defects in eggshell patterning (Morris and Lehmann 1999), these findings suggest that oogenesis in Top2-complementing females occurs without checkpoint activation. Eggs from Top235-5/Top235-13- and Top217-6/Top235-1-complementing females were collected and stained with DAPI. Imaging using confocal microscopy showed that the vast majority of eggs did not undergo nuclear divisions (90 of 95 and 30 of 31, respectively). While the lack of development may have resulted from a lack of fertilization, the observation that a few eggs displayed evidence of nuclear division suggests that fertilization could occur. We consider that compromised Top2 activity may lead to defects in transcription during oogenesis, causing depletion of maternal products needed in embryogenesis. Alternatively, these eggs may carry defects in meiosis that inhibit embryogenesis.
Fertility of complementing Top2 adults
| . | Allele transmitted by female (F) . | |||||
|---|---|---|---|---|---|---|
| Allele transmitted by male (M) . | 17-1 WHD . | 17-3 TOPRIM . | 17-6 TOPRIM . | 35-1 TOPRIM . | 35-5 ATPase . | 35-13 WHD . |
| 17-1 | NA | F NA | F NA | F sterile | F NA | NA |
| WHD | M fertile | M fertile | M fertile | M fertile | ||
| 17-3 | F NA | NA | F sterile | F sterile | F sterile | F sterile |
| TOPRIM | M fertile | M fertile | M sterile | M fertile | M fertile | |
| 17-6 | F sterile | F sterile | NA | F sterile | NA | F sterile |
| TOPRIM | M fertile | M fertile | M fertile | M fertile | ||
| 35-1 | F sterile | F sterile | F sterile | NA | F sterile | F sterile |
| TOPRIM | M fertile | M sterile | M fertile | M fertile | M fertile | |
| 35-5 | F NA | F sterile | NA | F sterile | NA | F sterile |
| ATPase | M fertile | M fertile | M fertile | M fertile | ||
| 35-13 | NA | F sterile | F sterile | F sterile | F sterile | NA |
| WHD | M fertile | M fertile | M fertile | M fertile | ||
| . | Allele transmitted by female (F) . | |||||
|---|---|---|---|---|---|---|
| Allele transmitted by male (M) . | 17-1 WHD . | 17-3 TOPRIM . | 17-6 TOPRIM . | 35-1 TOPRIM . | 35-5 ATPase . | 35-13 WHD . |
| 17-1 | NA | F NA | F NA | F sterile | F NA | NA |
| WHD | M fertile | M fertile | M fertile | M fertile | ||
| 17-3 | F NA | NA | F sterile | F sterile | F sterile | F sterile |
| TOPRIM | M fertile | M fertile | M sterile | M fertile | M fertile | |
| 17-6 | F sterile | F sterile | NA | F sterile | NA | F sterile |
| TOPRIM | M fertile | M fertile | M fertile | M fertile | ||
| 35-1 | F sterile | F sterile | F sterile | NA | F sterile | F sterile |
| TOPRIM | M fertile | M sterile | M fertile | M fertile | M fertile | |
| 35-5 | F NA | F sterile | NA | F sterile | NA | F sterile |
| ATPase | M fertile | M fertile | M fertile | M fertile | ||
| 35-13 | NA | F sterile | F sterile | F sterile | F sterile | NA |
| WHD | M fertile | M fertile | M fertile | M fertile | ||
NA, not assessed. Males were considered fertile if any progeny eclosed; females were considered fertile if they produced eggs that hatched.
| . | Allele transmitted by female (F) . | |||||
|---|---|---|---|---|---|---|
| Allele transmitted by male (M) . | 17-1 WHD . | 17-3 TOPRIM . | 17-6 TOPRIM . | 35-1 TOPRIM . | 35-5 ATPase . | 35-13 WHD . |
| 17-1 | NA | F NA | F NA | F sterile | F NA | NA |
| WHD | M fertile | M fertile | M fertile | M fertile | ||
| 17-3 | F NA | NA | F sterile | F sterile | F sterile | F sterile |
| TOPRIM | M fertile | M fertile | M sterile | M fertile | M fertile | |
| 17-6 | F sterile | F sterile | NA | F sterile | NA | F sterile |
| TOPRIM | M fertile | M fertile | M fertile | M fertile | ||
| 35-1 | F sterile | F sterile | F sterile | NA | F sterile | F sterile |
| TOPRIM | M fertile | M sterile | M fertile | M fertile | M fertile | |
| 35-5 | F NA | F sterile | NA | F sterile | NA | F sterile |
| ATPase | M fertile | M fertile | M fertile | M fertile | ||
| 35-13 | NA | F sterile | F sterile | F sterile | F sterile | NA |
| WHD | M fertile | M fertile | M fertile | M fertile | ||
| . | Allele transmitted by female (F) . | |||||
|---|---|---|---|---|---|---|
| Allele transmitted by male (M) . | 17-1 WHD . | 17-3 TOPRIM . | 17-6 TOPRIM . | 35-1 TOPRIM . | 35-5 ATPase . | 35-13 WHD . |
| 17-1 | NA | F NA | F NA | F sterile | F NA | NA |
| WHD | M fertile | M fertile | M fertile | M fertile | ||
| 17-3 | F NA | NA | F sterile | F sterile | F sterile | F sterile |
| TOPRIM | M fertile | M fertile | M sterile | M fertile | M fertile | |
| 17-6 | F sterile | F sterile | NA | F sterile | NA | F sterile |
| TOPRIM | M fertile | M fertile | M fertile | M fertile | ||
| 35-1 | F sterile | F sterile | F sterile | NA | F sterile | F sterile |
| TOPRIM | M fertile | M sterile | M fertile | M fertile | M fertile | |
| 35-5 | F NA | F sterile | NA | F sterile | NA | F sterile |
| ATPase | M fertile | M fertile | M fertile | M fertile | ||
| 35-13 | NA | F sterile | F sterile | F sterile | F sterile | NA |
| WHD | M fertile | M fertile | M fertile | M fertile | ||
NA, not assessed. Males were considered fertile if any progeny eclosed; females were considered fertile if they produced eggs that hatched.
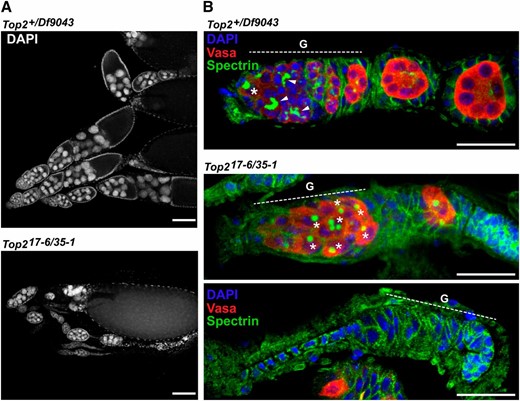
Low egg production prompted us to examine the ovary phenotype in Top217-6/Top235-1-complementing females. While strings of developing egg chambers were observed, the ovaries were smaller and disorganized and contained evidence of egg chamber apoptosis (Figure 6A). Effects of lowered Top2 function on the germline stem cell (GSC) niche were examined by staining with antibodies against Vasa, a germline-specific RNA helicase (Lasko and Ashburner 1988), and Spectrin, a structural protein expressed in a cell-type–specific pattern in all cells of the germarium. In germ cells, Spectrin accumulates in a spherical structure in the GSCs, called the spectrosome, and a branched structure in differentiating cysts, called the fusome, while in somatic cells, Spectrin localizes within the plasma membrane (Lin et al. 1994). We found that Top217-6/Top235-1 ovaries had complex defects in the germaria, including the presence of germaria showing both loss and gain of GSCs (Figure 6B). Taken together, these analyses indicate that germ cell differentiation is compromised in Top2-complementing females, leading to germ cell loss and defects in egg chamber formation.
Top2-complementing females show complex ovary phenotypes. (A) Wild-type and heteroallelic Top217-6/Top235-1 mutant ovaries were dissected from 3-day-old females and stained with DAPI. The ovaries of Top2-complementing females were smaller than wild-type ovaries, but still retained strings of developing egg chambers. Bars, 25 μm. (B) Wild-type and heteroallelic Top217-6/Top235-1 3-day-old ovaries were stained with DAPI and with antibodies against Vasa (red) to mark germ cells or with Spectrin (green) to mark spectrosomes (asterisks) present in germline stem cells and fusosomes (arrowheads) present in differentiating germ cells. Ovaries from the Top2-complementing females contained disorganized germaria (G), with a single ovary having germaria filled only with germ cells containing spectrosomes and germaria devoid of germ cells. Bars, 100 μm.
Concluding perspectives
Type II topoisomerases are molecular targets for chemotherapy against several types of cancers (Nitiss 2009b; Chikamori et al. 2010). However, treatment of cells with Top2 chemotherapeutic agents can produce drug-resistant forms of the enzyme. For example, treatment of bacteria, yeast, and human cells with chemotherapeutic agents has caused Top2 mutations that alter drug binding, DNA binding, or catalytic function (Nitiss 2009b; Wu et al. 2011). These drug-resistant forms of the protein commonly contain amino acid substitutions in the N-terminal ATPase and TOPRIM domains and the WHD. Interestingly, four of our Top2 missense alleles carry changes in amino acid residues previously found to be associated with drug resistance (Nitiss 2009b; Wu et al. 2011). These include Top217-3, Top235-1, Top235-6, and Top 35-12, mutations that have alterations in the TOPRIM domain and the WHD (Table 1) and correspond to residues L491, P485, S763, and G760 in human TopIIα, respectively. Among these mutations, Top217-3 and Top235-1 are hypomorphic alleles that support interallelic complementation, suggesting that certain heterodimers of drug-resistant proteins may similarly reconstitute enough Top2 function for viability. Based on these data, it is possible that individuals carry Top2 variants composed of defective monomers, which might confer different sensitivities to inhibitors and affect treatment outcomes.
Acknowlegments
We thank Donna Arndt-Jovin for the generous gift of Top2 antibodies. We thank Chantal Allamargot, Kathy Walters, and other staff at the Central Microscopy Research Facility at the University of Iowa for their assistance. We thank members of the Geyer and Wu laboratories for helpful discussions and comments on the manuscript and Anna Moran for technical assistance. This work was supported by National Institutes of Health grants to P. Geyer (GM42539) and C.-ting Wu (GM085169) and by a National Science Foundation Graduate Research Fellowship to A. Hohl.
Literature Cited
Footnotes
Communicating editor: J. A. Birchler
Author notes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.112.144006/-/DC1.
Present address: Center for Human Genetic Research, Massachusetts General Hospital, Boston, MA 02114.
Present address: Department of Pediatrics, Baylor College of Medicine, Houston, TX 77030.



![Structure of the Top2 locus. (A) Top2 is located on chromosome 2L between the uncharacterized upstream CG10026 gene and the essential downstream RanGap gene. Shown are the structures of three Top2 deficiency chromosomes used in these studies. Top2Df9043 is a 14.8-kb deletion allele (dashed line) that removes Top2, RanGap, Hs2st (the gene within the RanGap intron), and CG10237. Top2Df17 is an ∼3.6-kb deletion allele (dashed line) that removes only Top2 sequences and retains ∼600 bp of the starting P element (Top2EP). Top2Df35 is a deletion allele that removes Top2 sequences, but has unknown limits (dotted line). Promoters are indicated by bent arrows and exons are represented by shaded rectangles. (B) P[Top2-w+] is a P transposon that carries white+ (not shown), a 7.1-kb genomic fragment encompassing the entire Top2 gene, and the 5′ region of CG10026 and the 3′ largely untranslated region of RanGap. The coding region of the Top2 gene is annotated to indicate locations of the ATPase domain, the Transducer domain (TDD), the Topoisomerase/Primase (TOPRIM) domain, the Winged helix domain (WHD), the Tower domain (TD), the Coiled-coiled domain (CCD), and the carboxyl-terminal domain (CTD).](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/genetics/192/3/10.1534_genetics.112.144006/9/m_843fig1.jpeg?Expires=1716496199&Signature=uKhaEt~DpUZy0nw9FZ8z-56G5qmuquECTobvxJ54SzOIixLDlELpOMen~fZiwRm05ydOFNQ4-qxeraZ6rW8qKPZZM-G3Wx8SSVjrXKVfk9Bzzt2PbGZXzh~3vlpPfz~M64nIPPvYX24rzcypHKT2H-A6NSLwNi4O3dBPpMp4BExLPG~40axVkzDOI2xBvx1SUHUqp~bAj3TuDPsaYWbGaw4GpvetDlTgxp~DibOHd6geUcQOhP~7-he4VUdUpG-MCsDNBdf85OtwOhWoT6qhrXIhbQYX2Z2p7ATK4Vo3t44dVtWJOr9ZzRpQPyUdw8LWHROMnL8eS-cQY~nIkSVSjg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Loss of Top2 affects chromosome structure. Salivary gland polytene chromosomes were prepared from male third-instar larvae representing wild-type [Top2+/Top2Df9043 (A, D, and G)] or mutant [Top217-5/Top2Df9043 (B, E, and H) and Top235-14/Top2Df9043 (C, F, and I)] Top2 genotypes. Chromosomes were stained with DAPI (A–C), with enlargement of the male X chromosome shown to demonstrate the diffuse DNA-banding pattern (A′–C′). Recruitment of the MSL complex was assessed using antibodies specific to MSL-1 (D–F) and H4K16ac (G–I).](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/genetics/192/3/10.1534_genetics.112.144006/9/m_843fig5.jpeg?Expires=1716496200&Signature=4Vo-2c5ahiPF2-W0yDTROmU0fqFOqeStfe4~bJ2vzxRCqO2aeugiove-u1aV~OrS8LBDXn9igCqe2wLzzZbI4SGx1ayXQW81btccXfX0UxEbBP8MZ2qJN5XwFv55dfzcqvx5zoWjEAVwaLTQTdJ9z7eUA7RQ3kdsM7uhXtqxxFEMErTiTfSbXjbmf9CbD8rhCbzgQ22mx8lwRvsXPqJg71eXD2keQLXcW4gITPVDdjGhoW0IpcrGbJCz2HCs0CEtXQzeJuRJiZF0un5UkSwx-Ci0wpCpMs3M0aAQrMCSC8~95iVkQd5D4RyzcRYR5tDBnO8XSQmyer7RUTBNsQg5nQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)