-
PDF
- Split View
-
Views
-
Cite
Cite
Lei Xu, Janet Paulsen, Young Yoo, Elizabeth B Goodwin, Susan Strome, Caenorhabditis elegans MES-3 Is a Target of GLD-1 and Functions Epigenetically in Germline Development, Genetics, Volume 159, Issue 3, 1 November 2001, Pages 1007–1017, https://doi.org/10.1093/genetics/159.3.1007
Close - Share Icon Share
Abstract
The maternal-effect sterile (MES) proteins are maternally supplied regulators of germline development in Caenorhabditis elegans. In the hermaphrodite progeny from mes mutant mothers, the germline dies during larval development. On the basis of the similarities of MES-2 and MES-6 to known transcriptional regulators and on the basis of the effects of mes mutations on transgene expression in the germline, the MES proteins are predicted to be transcriptional repressors. One of the MES proteins, MES-3, is a novel protein with no recognizable motifs. In this article we show that MES-3 is localized in the nuclei of embryos and germ cells, consistent with its predicted role in transcriptional regulation. Its distribution in the germline and in early embryos does not depend on the wild-type functions of the other MES proteins. However, its nuclear localization in midstage embryos and its persistence in the primordial germ cells depend on wild-type MES-2 and MES-6. These results are consistent with biochemical data showing that MES-2, MES-3, and MES-6 associate in a complex in embryos. The distribution of MES-3 in the adult germline is regulated by the translational repressor GLD-1: MES-3 is absent from the region of the germline where GLD-1 is known to be present, MES-3 is overexpressed in the germline of gld-1 mutants, and GLD-1 specifically binds the mes-3 3′ untranslated region (3′ UTR). Analysis of temperature-shifted mes-3(bn21ts) worms and embryos indicates that MES-3 function is required in the mother's germline and during embryogenesis to ensure subsequent normal germline development. We propose that MES-3 acts epigenetically to induce a germline state that is inherited through both meiosis and mitosis and that is essential for survival of the germline.
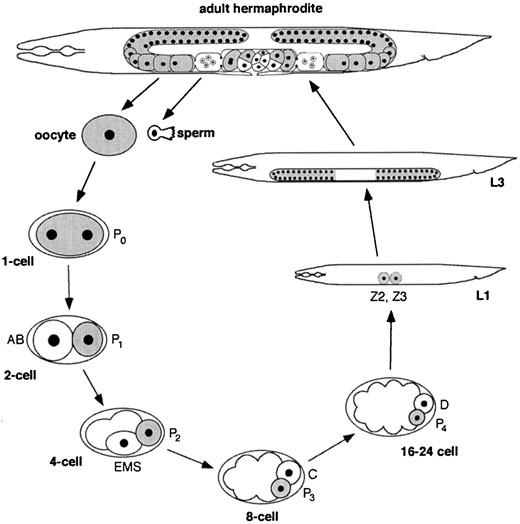
GERM cells are responsible for generating progeny, and, consequently, correct germline development is essential for the propagation of species. In recent decades, the nematode Caenorhabditis elegans has emerged as an excellent model system for studying germline development. It is fast growing, transparent, and has a prominent germline (Schedl 1997). C. elegans germline development can be observed continuously throughout the life cycle (Figure 1). After fertilization, the one-cell embryo undergoes four rounds of asymmetric division to produce the primordial germ cell, P4. During larval development the two daughters of P4, Z2 and Z3, undergo extensive proliferation to produce ~1500 germ cells in the two-armed gonad of adult hermaphrodites.
Each C. elegans gonad arm displays a distal-to-proximal polarity in its pattern of mitosis, meiosis, and gametogenesis (Schedl 1997). The most distal cells undergo mitotic proliferation and serve as a stem-cell population. More proximally, germ cells exit the mitotic cell cycle to enter and progress through prophase of meiosis I. Spermatogenesis occurs in the proximal gonad during the L4 stage. Hermaphrodites switch from spermatogenesis to oogenesis, which continues throughout adulthood.
Four maternal-effect sterile (mes) genes, mes-2, mes-3, mes-4, and mes-6, have been identified to be essential for germline development in C. elegans (Capowski et al. 1991). Homozygous mes progeny from heterozygous mothers are themselves fertile but produce sterile hermaphrodite progeny. The four mes genes are thought to be involved in a similar process, because their sterile phenotypes are similar: Sterility is caused by necrotic death of germ cells (Paulsen et al. 1995; Garvin et al. 1998). On the basis of their predicted protein sequences, MES-2 and MES-6 are the C. elegans orthologs of the Drosophila Polycomb group (PcG) proteins, Enhancer of Zeste [E(Z)] and Extra Sex Combs (ESC), respectively (Holdeman et al. 1998; Korf et al. 1998). MES-3 is a novel protein that does not contain any recognizable motifs (Paulsen et al. 1995). MES-4 contains a SET domain and multiple PHD fingers (Y. Fong, L. Bender and S. Strome, unpublished results).
PcG proteins are transcriptional repressors conserved in diverse species (Simon 1995; Gould 1997; Preuss 1999). The best known targets of PcG genes in Drosophila are homeotic genes, transcription factors whose expression
The germline cycle of C. elegans. The adult hermaphrodite has a two-armed gonad in which germ nuclei are in a syncytium. They divide mitotitically at the distal tip of each arm, enter meiosis, and differentiate into oocytes in the proximal arm. Oocytes are fertilized as they pass through the spermatheca and encounter sperm made during the L4 stage. The newly fertilized embryo (P0) undergoes four rounds of asymmetric divisions to form the primordial germ cell P4. P4 divides into Z2 and Z3, which proliferate during larval development to generate the ~1500 germ cells in the adult. Adapted from Strome et al. (1995).
patterns determine the anterior-posterior body pattern (Gould 1997; Pirotta 1998). The expression pattern of homeotic genes is established initially by transiently expressed gap and pair-rule proteins in early embryos. At later stages, this expression pattern is maintained by the antagonistic functions of two groups of proteins, PcG and trxG (trithorax group) proteins (Simon 1995; Pirrotta 1998). PcG proteins maintain repression of homeotic genes outside of their expression domains, and trxG proteins maintain active expression within their expression domains. Repression by PcG proteins is mediated by chromosomal elements called PREs (PcG response elements; Simon et al. 1993; Chan et al. 1994; Gindhardt and Kaufman 1995; Horard et al. 2000). PcG proteins bind to PREs in vivo and package the adjacent chromatin into a closed repressed conformation.
Recently, Cavalli and Paro (1998) presented evidence that the repressed chromosomal state induced by PcG proteins is heritable through cell divisions. They used Drosophila transgenic lines carrying a GAL4-driven lacZ-miniwhite reporter flanked by the Fab-7 element. Fab-7 is a PRE-containing cis-regulatory element in the bithorax complex (Busturia and Bienz 1993) that can confer PcG-mediated silencing of adjacent transgenes. PcG-mediated silencing via Fab-7 is enhanced at high temperatures (28°; Fauvarque and Dura 1993; Zink and Paro 1995). When transgenic animals containing the Fab-7 PRE near a GAL4-inducible lacZ-miniwhite reporter were shifted to 28° during embryogenesis and subsequently reared at 18°, the hyper-repressed state of the reporter genes induced during embryogenesis was maintained throughout the rest of their development. This result suggests that PcG-mediated silencing through Fab-7 is mitotically heritable. The results of experiments done to address meiotic transmission of the chromatin state further suggest that PcG-mediated repression via Fab-7 can be transmitted through meiosis to the next generation at a certain frequency.
MES proteins are predicted to repress gene expression in the C. elegans germline, as PcG proteins do in the Drosophila soma. This prediction is supported mainly by analysis of transgene expression in the germline of C. elegans (Kelly and Fire 1998). Transgenes injected into C. elegans gonads recombine with each other and form extrachromosomal arrays. They are usually expressed in somatic cells but are silenced in the germline (Kelly et al. 1997). This silencing is thought to be a consequence of the repetitive nature of extrachromosomal arrays. Transgenes can be desilenced in the germline by generating arrays containing complex genomic DNA and a reduced copy number of transgenes (Kelly et al. 1997). Interestingly, transgenes in repetitive arrays are desilenced in the germlines of mes mutants, indicating that in wild-type germlines MES proteins participate in repressing transgene expression from repetitive arrays (Kelly and Fire 1998).
Here we present molecular analysis of one of the MES proteins, MES-3. It is localized in the nuclei of embryos and of germ cells. Molecular epistasis results show that the correct distribution of MES-3 depends upon MES-2 and MES-6 during a midstage of embryogenesis but not in early embryos or in the adult germline. Temperature-shift studies of a temperature-sensitive allele of mes-3 show that MES-3 function is required in the maternal germline and during embryogenesis for subsequent normal germline development. These data suggest that regulation by MES proteins in C. elegans is epigenetically heritable, which is similar to regulation by PcG proteins in Drosophila.
MATERIALS AND METHODS
Alleles and strain maintenance: C. elegans N2 variety Bristol is the wild-type strain used in this article. The mes-3 alleles bn21ts, bn35, and bn53 were induced by EMS mutagenesis; bn86 and bn88 were induced by gamma radiation (Paulsen et al. 1995). The following mutations, duplications, and balancers were used:
LGI: mes-3(bn21, bn35, bn53, bn86, bn88), dpy-5(e61), hDp20(I;V, f)
LGII: mes-2(bn11), unc-4(e120), mnC1[dpy-10(e128) unc-52 (e444)]
LGIV: mes-6(bn38), nT1[let(m435)](IV;V), DnT1[unc (n754 dm) let](IV;V)
LGV: mes-4(bn67), dpy-11(e224), nT1[let(m435)](IV;V), DnT1 [unc(n754dm) let](IV;V)
Strains were maintained following standard procedures (Brenner 1974).
Sequence analysis of mes-3 alleles: All five alleles were sequenced using an ABI (Columbia, MD) PRISM DNA sequencing kit and ABI PRISM 310 genetic analyzer (PE Applied Biosystems). Genomic DNA was prepared from homozygous mes-3 mutants carrying each allele, and the mes-3 gene was amplified by PCR using the following eight sets of 5′ and 3′ primers: set 1 (5′-CTGATTGGCGATTTCGAG-3′ and 5′-GTTTGTCGAAGACATTACG-3′), set 2 (5′-TCAGCTCGTCGAGAAGAAG-3′ and 5′-GTCGACAATTATCACGTAGC-3′), set 3 (5′-AGCCAGAAAACGCGAAAATG-3′ and 5′-TACGCTGGCTTTTCTTCTCG-3′), set 4 (5′-CCAAAATTATCCGAAGGTG-3′ and 5′-GCGAATATCCAATGTAGTG-3′), set 5 (5′-ATCGCAATATGAAGAGTTCG-3′ and 5′-TCTCAGGAACTCAATATCTG-3′), set 6 (5′-CCAGTTTCCGTTCCAATTG-3′ and 5′-GTGGATAGTCAAGAATTGTC-3′), set 7 (5′-TGTGACATGTTGCTCCAAC-3′ and 5′-AGAGATTAAACACAGGATTTC-3′), and set 8 (5′-ACCATCAGTACATTTTCGAC-3′ and 5′-ATGGCATCCTTGATTGGAG-3′). Each fragment was sequenced using the same primers that were used for PCR. Sequencing results were compared with the wild-type mes-3 gene sequence in GenBank using the Sequencher 3.0 program. Mutations in each allele were verified by repeating the PCR and sequencing reactions.
Antibody production: A C-terminal peptide of MES-3, CRYK QLKKEWKNRQRDIPSSN (the Cys was added for conjugation), was synthesized and conjugated to the KLH carrier protein by Research Genetics (Huntsville, AL), and rat antisera were produced by Cocalico Biologicals. Anti-MES-3 antibodies were affinity purified against a (His)6-tagged MES-3 fusion protein, which contains the C-terminal 272 amino acids of MES-3 and was conjugated to an affi-gel 10 (Bio-Rad, Richmond, CA) column. The purified antibodies were eluted from the column with 0.2 m Gly-HCl pH 2.5.
Western blot analysis: To test the specificity of anti-MES-3 antibody, ~50 N2 or mes-3 (bn35) homozygous worms were picked into 1× SDS sample buffer and boiled for 5 min. The proteins from lysed worms were separated on an 8% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. To probe the membrane, affinity-purified rat anti-MES-3 antibody (1:50) or mouse anti-α-tubulin (1:200) was used as the primary antibody, and horseradish peroxidase-conjugated goat anti-rat antibody (1:2000) or goat anti-mouse antibody (1:2000; Jackson Laboratories) was used as the secondary antibody. The signal was developed using an enhanced chemiluminescence kit (Amersham Pharmacia Biotech).
To determine whether the level of MES-3 is reduced in mes-3 (bn21ts) mutants, N2 and mes-3(bn21) homozygous worms were raised at 16° and 25° from the L1 stage onward. Fifty of the resulting adults were picked and boiled in 1× SDS sample buffer for Western blot analysis as described above.
Immunostaining: Worms were cut open, fixed by methanol and acetone followed by air drying, and stained as previously described (Strome and Wood 1983). L1s were not cut but were otherwise fixed and stained in the same way. Affinity-purified rat anti-MES-3 antibody (1:5) was used as the primary antibody, followed by Alexa 488-conjugated goat anti-rat secondary antibody (1:200; Molecular Probes, Eugene, OR). 4′,6-Diamidino-2-phenylindole (DAPI) was used to visualize DNA. Samples were mounted in gelutol (DuPont, Wilmington, DE). The staining patterns described were reproducible in at least 50 worms. Immunofluorescence images were captured and processed as previously described (Kawasaki et al. 1998). When comparing the staining pattern between N2 and mes mutants, or between N2 and gld-1 mutants, all staining incubations were performed side-by-side following the same procedures. All images were captured on Kodak Tri-X 100 pan film for the same exposure time, scanned using a SprintScan35 (Polaroid), and processed identically using Adobe Photoshop.
For confocal microscopy, worms were cut, fixed by methanol and acetone, and stored in PBS (without air drying) before being stained. The rest of the staining procedure was as described above. Rabbit anti-penta-acetylated histone H4 antibodies (1:1000; Upstate Biotechnology, Lake Placid, NY; Lin et al. 1989) were used to visualize chromosomes. Images were captured as a stacked series by a Bio-Rad MRC 600 confocal microscope and processed using NIH Image (National Institutes of Health) and Adobe Photoshop.
GLD-1 gel-shift analysis: RNA gel shift was performed as described (Goodwin et al. 1993). 32P-labeled RNA probes containing the mes-3 wild-type 3′ untranslated region (UTR), or unlabeled RNA probes containing either the tra-2 wild-type 3′ UTR or a mutant tra-2 3′ UTR in which the TGEs (tra-2 and GLI elements) were deleted, were made by standard methods using the RiboMAX kit (Promega, Madison, WI). The different full-length and mutant 3′ UTRs were subcloned into KSII(+) pBluescript vector. The 3′ UTR-containing pBluescript vectors were linearized and the sense 3′ UTR RNAs were transcribed in vitro by either T3 or T7. Quantitation of the cold RNA probes was measured by spectrophotometry.
Temperature-shift experiments: Embryonic temperature-shift experiments with mes-3 (bn21ts) were performed as follows. For upshift experiments (16° to 25°), embryos were isolated from gravid hermaphrodites that had been raised at the permissive temperature (16°). Gravid hermaphrodites were placed in a drop of M9 buffer (Brenner 1974) on a gelatin-coated slide and cut open to expel the embryos. Embryos were examined on a transmission dissecting microscope at ×100 magnification. One-, two-, four-cell, and comma-stage embryos, which are easily distinguished, were isolated and transferred to a new drop of M9 and placed at 25°. The embryos were transferred to 25° plates with food 1–4 hr later. To obtain embryos between the four-cell and comma stage, four-cell embryos were isolated and kept at 16° for various amounts of time before being transferred to 25°. To determine the stage of the embryos at the time of their transfer from 16° to 25°, 5–15 embryos from the same time points were fixed and immunostained with anti-P-granule antibodies and DAPI, as described previously, and the nuclei and P-granule-containing cells were counted. L1 larvae were obtained by placing mixed-stage embryos in a drop of M9 and allowing them to hatch at 16°. The newly hatched L1 larvae were then transferred to the restrictive temperature. In 2–3 days, the experimental embryos and larvae (between 31 and 45 for each time point) were scored as adults for sterility vs. fertility.
For downshift experiments, embryos were isolated from gravid hermaphrodites that had been raised for part of larval development at the restrictive temperature (25°). In the downshift a experiment, mother worms were transferred from 16° to 25° as L1 to L2 larvae. In the downshift b experiment, mother worms were transferred from 16° to 25° as L3 to early L4 larvae. After these mothers became gravid, embryos and larvae (between 26 and 51 for each time point) were obtained at 25°, shifted to 16° using the procedures described above, grown to adulthood, and scored for sterility vs. fertility.
RESULTS
Sequence analysis of mes-3 alleles: mes-3 was previously reported to be the upstream gene of an operon and to encode a novel protein with no recognizable motifs (Paulsen et al. 1995). To investigate what portion of
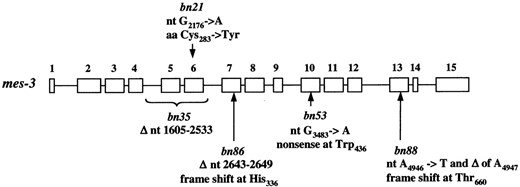
Mutations in mes-3 alleles. The mes-3 gene is schematically shown as 15 exons (open boxes) and 14 introns (lines between boxes). The positions of mutations (position 1 is the nucleotide adjacent to the trans-spliced leader) in each of the five alleles of mes-3 are indicated by a bracket (for bn35) or arrows. ▵, deletion; nt, mutation in the nucleotide sequence of mes-3; aa, amino acid alteration in MES-3. Our sequencing confirmed that in the wild-type gene, aa264 is Ala, as shown for F54C1.3 in GenBank.
MES-3 protein is important for its function, we sequenced the five alleles of mes-3 (Figure 2). One allele, bn21, is temperature sensitive: bn21 mutants produce fertile progeny at permissive temperature (16°) and sterile progeny at restrictive temperature (25°). The bn21 allele has a missense mutation of Cys283 → Tyr. The other four alleles, which are nonconditional, contain mutations that create premature stop codons. bn35 has a 929-bp deletion that removes exons 5 and 6 and parts of the upstream and downstream introns; splicing of exon 4 to exon 7 would result in a frameshift and a premature stop codon in exon 7. bn86 contains a 7-bp deletion in exon 7, which also causes a frameshift and a premature stop codon right after the deletion. bn53 has a point mutation, which changes Trp436 to a stop codon in exon 10. bn88 contains a point mutation followed by a single nucleotide deletion in exon 13, which creates a frameshift and a premature stop codon in exon 14.
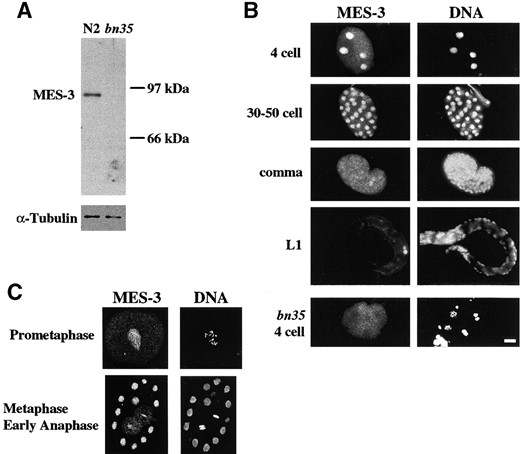
MES-3 protein is localized in the nuclei of embryos: To gain insights into where and when MES-3 protein functions, antibodies against the C-terminal peptide of MES-3 were produced and the distribution of MES-3 was determined by immunostaining. We found that MES-3 is localized predominantly in nuclei (Figure 3B). In early embryos, MES-3 protein is present in the nuclei of all cells. As embryogenesis progresses, staining gradually diminishes in somatic cells. In late embryos and L1 larvae, MES-3 is detectable in some somatic cells but is most prominent in Z2 and Z3, the primordial germ cells. The nuclear staining of MES-3 is reduced below detection in any of the four nonconditional alleles of mes-3. This result and antibody recognition of a worm protein of the predicted size (~90 kD) in N2 lysate, but not in mes-3(bn35) lysate on Western blots (Figure 3A), demonstrate the specificity of the antibodies.
To visualize whether MES-3 protein is localized on chromosomes, we analyzed the immunolocalization pattern of MES-3 in embryos, using confocal microscopy (Figure 3C). Cells at different stages of mitosis were stained by affinity-purified anti-MES-3 antibody and antipenta-acetylated H4 antibody to visualize chromosomes. During interphase (data not shown) and prometaphase (Figure 3C), when condensed chromosomes are clearly visible in nuclei, MES-3 protein is not obviously concentrated on chromosomes; instead it appears evenly distributed in the nucleoplasm. During metaphase and early anaphase, when nuclear envelopes are broken down, some MES-3 protein is detectably associated with chromosomes (Figure 3C).
Western blot and immunostaining of embryos by rat anti-MES-3 antibodies. (A) Affinity-purified rat anti-MES-3 antibody was used to probe wild-type (N2) or mes-3 (bn35) worm extract that had been size fractionated by SDS-PAGE and blotted to nitrocellulose membrane. One band of ~90 kD was recognized by the anti-MES-3 antibody in the N2 lane but not in the bn35 lane. The same blot was probed with mouse anti-α-tubulin antibody as a loading control (bottom). (B) Wild-type or mes-3(bn35) embryos were stained with affinity-purified rat anti-MES-3 antibody and DAPI to visualize DNA. The stage of the embryo is indicated on the left. Four-cell embryo, staining is nuclear in all four cells; 30–50-cell embryo, MES-3 is present in all nuclei; Comma-stage embryo, faint staining is seen in all nuclei. The primordial germ cells Z2 and Z3 (~one-third from right end of embryo) are more brightly stained. L1 larva, staining is detected primarily in the primordial germ cells Z2 and Z3 (right); bn35 4-cell embryo, nuclear staining is not detectable. Bar, 10 μm. (C) Confocal images of wild-type embryos stained with rat anti-MES-3 and rabbit anti-histone to visualize chromosomes. Prometaphase, MES-3 is distributed in the nucleoplasm; metaphase and early anaphase, a portion of MES-3 is localized on chromosomes.
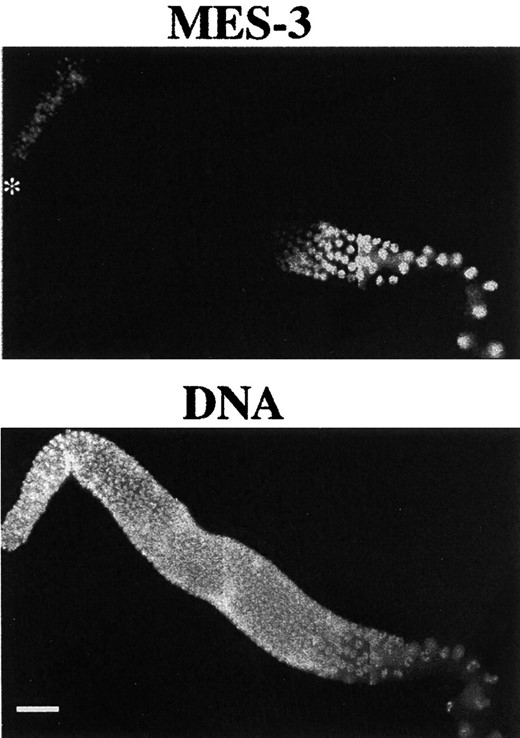
MES-3 localization in the germline. Gonad arms dissected from wild-type hermaphrodites were stained with affinity-purified rat anti-MES-3 and DAPI. The distal tip is indicated by an asterisk. Oocytes are in the bottom right corner. MES-3 is observed in the nuclei of the distal mitotic region, the proximal meiotic region, and oocytes. Its level is reduced to below detection in nuclei in early pachytene. Bar, 20 μm.
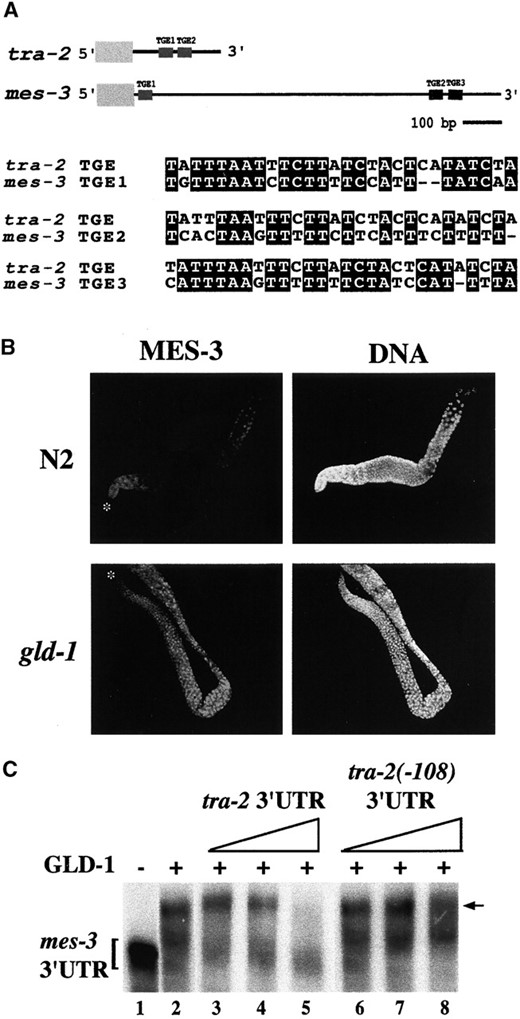
The distribution of nuclear MES-3 is regulated by GLD-1 in the adult germline: In adults, MES-3 is most prominent in germline nuclei (Figure 4) and is occasionally barely detectable in intestinal nuclei (data not shown). In the germline, it is present at low levels in distal mitotic nuclei, undetectable in the pachytene region of the distal arm, and present at elevated levels in the proximal meiotic region and in oocytes (Figure 4). This pattern led us to speculate that MES-3 expression might be repressed in the pachytene region of the germline. Several germline genes, such as fem-3 (Ahringer and Kimble 1991), tra-2 (Goodwin et al. 1993; Jan et al. 1999), and glp-1 (Evans et al. 1994), have been found to be regulated post-transcriptionally via their 3′ UTRs. In fact, the 3′ UTR of the mes-3 transcript contains TGE-like sequences (Figure 5A). TGEs are the translational regulatory elements originally found in the 3′ UTR of tra-2 (Jan et al. 1997), where they are present as two consecutive repeats. The mes-3 3′ UTR contains three TGE-like sequences, one in the 5′ region of the 3′ UTR and two consecutive motifs in the 3′ region of the 3′ UTR (Figure 5A).
GLD-1, an RNA-binding protein, binds to the TGEs in the tra-2 mRNA and inhibits its translation (Jan et al. 1999). Interestingly, GLD-1 is expressed in the cytoplasm of pachytene-stage germ cells (Jones et al. 1996). Its level is significantly lower in the distal mitotic germ cells and in oocytes. This distribution pattern is complementary to the localization pattern of MES-3, raising the possibility that GLD-1 is responsible for repression of MES-3 translation in the pachytene region of the germline. We therefore examined the expression profile of MES-3 in the germline of gld-1 mutants (Figure 5B). The level of MES-3 appeared uniform throughout the germline of gld-1 mutants and appeared to be present at higher levels in gld-1 germlines than in wild-type germlines. These results suggest that GLD-1 represses MES-3 expression in the pachytene region of the adult germline, probably by binding to the TGE-like sequences in its 3′ UTR and by inhibiting its translation.
To further test whether GLD-1 may inhibit mes-3 mRNA activity, we performed RNA gel-shift analysis and asked if GLD-1 can specifically bind the mes-3 3′ UTR. Incubation of bacterially expressed purified GST-GLD-1 fusion protein with labeled RNA containing the mes-3 3′ UTR resulted in the appearance of a slower-migrating complex (Figure 5C; lane 2). To test if GLD-1 binding is specific to TGEs, an excess of either cold tra-2 wild-type 3′ UTR (Figure 5C; lane 3-5) or mutant tra-2 3′ UTR that lacks the TGEs (Figure 5C; lanes 6–8) was added to the reaction. An excess of the wild-type tra-2 3′ UTR, but not the mutant 3′ UTR, inhibited the interaction of the mes-3 3′ UTR with GLD-1. These findings indicate that GLD-1 can specifically bind the mes-3 3′ UTR in a TGE-dependent manner and are consistent with GLD-1 inhibiting mes-3 translation in the germline.
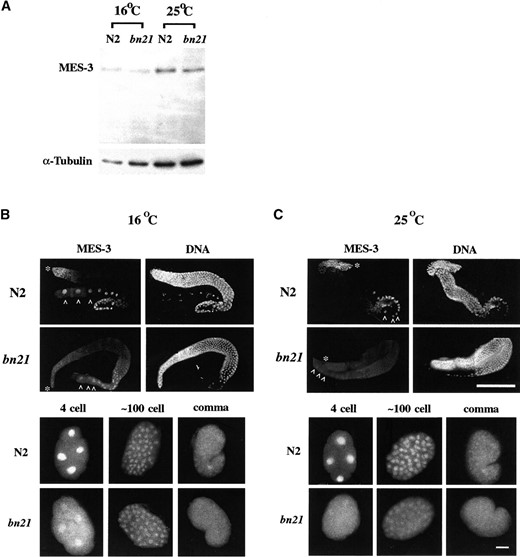
MES-3 protein is mislocalized, but not destabilized, in mes-3(bn21ts) mutants: To investigate whether the temperature sensitivity of mes-3(bn21) is due to destabilization of MES-3, we performed Western blot analysis of MES-3 protein levels in wild-type or mes-3(bn21) adults grown at 16° or 25° from the L1 stage. Embryos from bn21 worms raised at 16° grow up to be fertile, whereas embryos from those raised at 25° grow up to be sterile (see below). At both temperatures, the level of MES-3 in bn21 adults appears to be similar to the level in wild-type adults (Figure 6A), indicating that the sterility of bn21 mutants at 25° is not due to destabilization of the maternal load of MES-3.
To determine whether MES-3 is mislocalized in mes-3 (bn21) mutants, we immunostained wild-type and mes-3 (bn21) worms grown as described above with anti-MES-3 antibodies. At 16°, the MES-3 protein distribution in bn21 mutant germlines and embryos generally resembles that in wild type, although a greater proportion of MES-3 appears to be cytoplasmic in oocytes and early embryos (Figure 6B). At 25°, MES-3 protein appears to be predominantly cytoplasmic, both in the germline and in embryos (Figure 6C). These results show that the distribution of mutant MES-3 protein (containing Tyr instead of Cys at amino acid 283) is mildly defective at the permissive temperature and severely defective at the restrictive temperature. The defects at the restrictive temperature correlate with sterility.
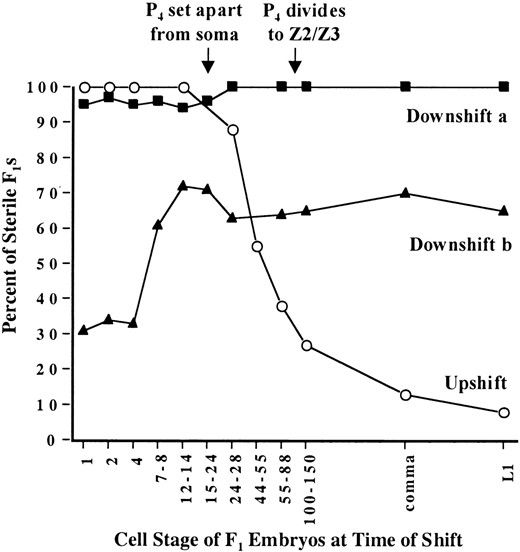
The temperature-sensitive period for mes-3(bn21ts) is during maternal germline development and during embryogenesis: To determine the temporal requirement for MES-3 protein, we analyzed the temperature-sensitive period (TSP) of mes-3(bn21) mutants. mes-3(bn21) embryos (F1 generation) were shifted from 16° to 25° or vice versa at different developmental stages, grown to adulthood, and scored for sterility vs. fertility (Figure 7). Upshifting bn21 embryos at any stage before the ~14-cell stage caused all of the shifted embryos to develop into sterile adults (Figure 7, upshift). Progressively later upshifts resulted in progressively fewer sterile adults. Almost all embryos upshifted at the L1 stage (Figure 7, upshift) or later (data not shown) developed into fertile adults. These results suggest that MES-3 function is not needed continuously during germline development in the F1 worms analyzed. Instead, our finding that the TSP of mes-3 ends between the 14-cell stage and the end of embryogenesis suggests that MES-3 function
mes-3 is a target of GLD-1 regulation. (A) The mes-3 3′ UTR contains three TGE-like sequences, which are aligned with the TGE sequence that is present twice in the 3′ UTR of tra-2. The alignments were done by NCSA BIOLOGY WORKBENCH software (http://workbench.sdsc.edu). Identical nucleotides are highlighted with black boxes. (B) Extruded gonads from wild-type (N2) and gld-1 worms were stained with affinity-purified rat anti-MES-3 and DAPI. The distal tips are indicated by asterisks (*). Oocytes in wild type are in the top right. gld-1 mutants do not make oocytes. In contrast to the dynamic distribution in wild type, MES-3 is evenly distributed throughout the germline of gld-1 mutants. The level of MES-3 appears higher in the germline of gld-1 mutants than in wild type. (C) Binding of GLD-1 to the mes-3 3′ UTR was tested using RNA gel mobility shift assay. (Lane 1) A 5-fmol aliquot of 32P-labeled mes-3 3′ UTR was incubated alone or (lanes 2–8) with 0.25 μg of GST-GLD-1 protein, which resulted in the appearance of a slower-migrating complex (arrow). To determine the specificity of GLD-1 binding, increasing amounts of cold wild-type tra-2 3′ UTR (lanes 3–5) or cold mutant tra-2 3′ UTR in which the TGEs had been deleted (lanes 6–8) were added to the reaction. The amounts of cold RNAs added to the reactions were 50, 250, and 500 fmol.
during and perhaps before embryogenesis is sufficient to ensure fertility.
To perform embryonic downshift experiments, it was necessary to culture the mother worms (P0 generation) at 25°. Interestingly, the time at which mothers (P0) were placed at 25° affected whether or not the fertility of their progeny (F1) could be rescued by shifting the F1's down to 16°. When mes-3(bn21) P0 mothers were placed at 25° as L1 or L2 larvae, downshifting their F1 progeny at any stage did not rescue fertility (Figure 7, downshift a). In contrast, when mes-3(bn21) P0 mothers were placed at 25° as L3 or early L4 larvae, downshifting their F1 progeny as very early embryos rescued fertility in ~70% of the F1's, and downshifting their F1 progeny at or after the 14-cell stage rescued fertility in ~30% of the F1's (Figure 7, downshift b). These downshift results support a model in which MES-3 function is required both in the mother's germline (P0) and in embryos (F1) for those embryos to develop into fertile adults (see discussion). A lack of MES-3 function in the maternal germline throughout all stages of larval development of the mother (P0) resulted in irreversible sterility of the F1's (downshift a), whereas the presence of normal MES-3 function in the maternal germline during the L1 and L2 stages partially rescued the fertility of the F1's (downshift b). In the latter case, the degree of rescue depended on when the F1 embryos were downshifted. Thus, both upshift and downshift results point to MES-3 functioning early in the F1 embryos for their normal germline development later on.
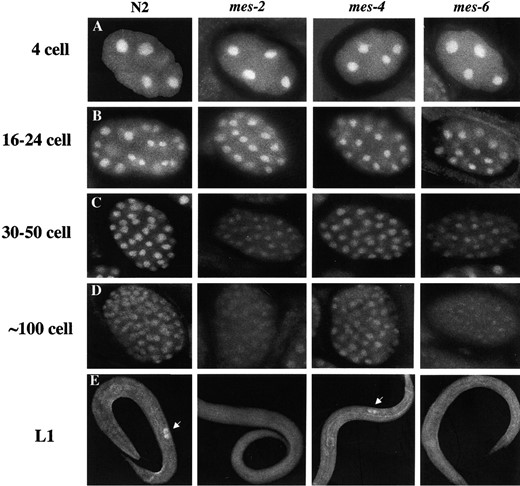
The distribution of MES-3 is altered in mes-2 and mes-6 mutants: To study the epistasis relationship between MES-3 and the other MES proteins, we performed immunostaining of MES-3 in other mes mutants (Figure 8). In the germlines of homozygous mes-2, mes-4, and mes-6 hermaphrodites from heterozygous mothers, the
Accumulation and distribution of MES-3 in mes-3(bn21) mutants. (A) Worm lysates from wild-type (N2) worms or mes-3(bn21) mutants raised at 16° or 25° were probed by affinity-purified anti-MES-3 antibody on a Western blot (top). The same blot was probed by mouse anti-tubulin antibody as a loading control (bottom). MES-3 accumulates to similar levels in N2 and mes-3 (bn21) mutants at both 16° and 25°. (The MES-3 bands are dimmer in the 16° lanes because the 16° worms were younger and contained fewer embryos than the 25° worms.) (B and C) N2 or mes-3(bn21) gonads and embryos were extruded from hermaphrodites grown at 16° or 25° and stained by affinity-purified rat anti-MES-3 antibody and DAPI. The distal ends of the gonads are indicated by asterisks (*) and the oocytes are indicated by white carets (∧). The images were captured by Kodak Tri-X pan film with a set exposure time, and pictures were processed identically afterward. (B) At 16°, the distribution of MES-3 in bn21 gonads and bn21 embryos is similar to the distribution in wild type, but a higher proportion of MES-3 is distributed in the cytoplasm in bn21 oocytes and early embryos than in wild type. (C) At 25°, the level of MES-3 in the germline appears reduced and more cytoplasmic in bn21 than in wild type. In embryos, most MES-3 protein is distributed in the cytoplasm. Bar for gonads, 100 μm; bar for embryos, 10 μm.
level and distribution of MES-3 appeared normal (data not shown). The embryos from these mes mutants, which develop into sterile adults, showed normal MES-3 patterns up to the 24-cell stage. However, at later stages, the level of MES-3 in nuclei gradually diminished in mes-2 and mes-6 mutant embryos but not in mes-4 mutant embryos. By the time of hatching, MES-3 was detected in the nuclei of the primordial germ cells, Z2 and Z3, in wild type and in mes-4 L1s, but was undetectable in those cells in mes-2 and mes-6 L1s. We do not know if the drop in nuclear MES-3 levels in late-stage mes-2 and mes-6 embryos is due to destabilization or mislocalization of MES-3. These results indicate that in >24-cell-stage embryos, a normal distribution of MES-3 depends on the presence of MES-2 and MES-6 but not on MES-4.
DISCUSSION
MES-3 expression is repressed by GLD-1 in the germline: Post-transcriptional control of the expression of MES-3 appears to occur in the adult hermaphrodite germline. Immunostaining analysis showed that MES-3 is not distributed evenly in wild-type germlines; instead, MES-3 is present in the nuclei of the distal mitotic and proximal meiotic regions but is undetectable in germ nuclei at the early pachytene stage. We think that this decrease in MES-3 level in the pachytene region is due to translational repression by GLD-1. The reasons are as follows:
In situ hybridization analysis by the Y. Kohara laboratory has shown that mes-3 mRNA is fairly evenly distributed throughout the germline (http://nematode.lab.nig.ac.jp). This result suggests that the decrease in MES-3 protein level is likely due to post-transcriptional regulation.
GLD-1 is present in the right region to be a MES-3 repressor. It is localized in the cytoplasm of pachytene cells, and its level is significantly lower in the mitotic region and in oocytes (Jones et al. 1996). Thus, it accumulates in the region where MES-3 levels are lowest.
mes-3 mRNA contains potential target sequences of GLD-1 and is specifically bound by GLD-1. Previously, GLD-1 was found to inhibit tra-2 mRNA translation by binding to TGEs in its 3′ UTR (Jan et al. 1999). The 3′ UTR of mes-3 mRNA contains TGE-like sequences similar to those in tra-2.
MES-3 protein is expressed uniformly and at a significantly higher level throughout the germline of gld-1 mutants, strongly suggesting that GLD-1 represses MES-3 expression in wild-type germlines.
TSP analysis for mes-3(bn21ts). Upshift data are graphed with open circles. Mothers (P0) were grown at 16°, and their embryos (F1) were shifted from 16° to 25° at different stages, grown to adulthood at 25°, and scored for sterility vs. fertility. Downshift a data are graphed with solid squares. Mothers were placed at 25° as L1 to L2 larvae; their embryos were shifted from 25° to 16° at different stages, grown to adulthood at 16°, and scored for sterility vs. fertility. Downshift b data are graphed with solid triangles. Mothers were placed at 25° as L3 to early L4 larvae; their embryos were shifted from 25° to 16° at different stages, grown to adulthood at 16°, and scored for sterility vs. fertility. See materials and methods for experimental details.
GLD-1 is an RNA-binding protein required for germ cells to complete meiosis and differentiate into oocytes (Jones et al. 1996). In gld-1 mutants, germ cells enter meiosis but then exit the meiotic pathway and return to mitotic cell cycles. As a consequence, gld-1 mutants present a tumorous mitotic germline phenotype. In addition to its essential role in oogenesis, previous genetic results suggest that gld-1 serves nonessential inhibitory roles in germline proliferation (Francis et al. 1995). GLP-1, a Notch homolog, is the germline receptor of a somatic signal for mitotic proliferation of germ cells. The germline proliferation defect in glp-1 mutants is suppressed partially in a gld-1(null) background. This suppression might be due to the overexpression of MES-3, which is essential for germline proliferation, in gld-1 mutants. This prediction may be tested by examining the phenotype of glp-1 mutants that overexpress MES-3 in their germlines. If the prediction is correct, then the germline proliferation defect created by glp-1 mutations should be rescued at least partially by overexpressed MES-3.
MES-2, MES-3, and MES-6 appear to associate in a complex in a stage-dependent manner: Epistasis analysis showed that MES-3 depends on wild-type MES-2 and MES-6 for its normal localization in late-stage embryos. MES-3 localization does not depend on MES-4. This is consistent with our previous observation that the localization of MES-2 and MES-6 depends on wild-type MES-3 but not on MES-4 (Holdeman et al. 1998; Korf et al. 1998). Taken together, these data suggest strongly that MES-2, MES-3, and MES-6 form and function as a complex and that MES-4 functions independently. In fact, the results of biochemical analyses have shown that MES-2, MES-3, and MES-6 are in a complex in embryo extracts and that MES-4 is not in this complex (Xu et al. 2001). This is similar to PcG and trxG proteins in Drosophila. Proteins from both groups form heterogeneous complexes to repress or activate gene expression (Papoulas et al. 1998; Shao et al. 1999; Ng et al. 2000).
Interestingly, we found that the dependence of MES-3 localization on MES-2 and MES-6 is stage specific in embryos. In early embryos (<30 cells), MES-3 localization appears normal in mes-2 and mes-6 mutants, indicating that MES-3 localization does not depend on MES-2 and MES-6 during early embryogenesis. In contrast, in these early-stage embryos, the nuclear localization of MES-2 and MES-6 depends on MES-3 (Holdeman et al. 1998; Korf et al. 1998). These data suggest that in early embryos MES-3 acts upstream of MES-2 and MES-6. MES-3 might enter nuclei and associate with chromosomal target sites before MES-2 and MES-6 and then recruit MES-2 and MES-6 to those sites and thereby stabilize MES-2 and MES-6 in nuclei. In later-stage embryos, MES-3 levels are reduced in mes-2 and mes-6 mutants compared to wild type. Conversely, in the absence of MES-3, MES-2 and MES-6 are not detectable in the nuclei of late embryos. These results suggest that in late embryos, an intact MES-2/MES-3/MES-6 complex is essential for the stable nuclear localization of each of the three MES proteins.
In the adult germline, MES-3 localization does not depend on MES-2 and MES-6, and the nuclear localization of MES-2 and MES-6 does not depend on MES-3 (Holdeman et al. 1998; Korf et al. 1998). These findings suggest that MES-3 does not associate with MES-2 and MES-6 in the germline. MES-2 and MES-6, however, depend upon each other for proper nuclear localization and thus probably interact with each other in the germline. The MES-2/MES-6 complex may function as an independent unit or may have a partner different from MES-3 in the germline. MES-4 is not required for the
Distribution of MES-3 in other mes mutants. Embryos and L1 larvae from wild-type (N2) or homozygous mes-2, mes-4, and mes-6 mothers were stained with affinity-purified rat anti-MES-3 antibody and DAPI (DNA not shown). The images were captured by Kodak Tri-100 pan film with a set exposure time, and pictures were processed identically afterward. In early embryos (4–24 cells), the nuclear localization of MES-3 appears similar in mes mutants and in wild type. In later-stage embryos (>30 cells), the level of MES-3 is similar in mes-4 mutants and in N2 but appears significantly reduced in mes-2 and mes-6 mutants. In L1 larvae, MES-3 protein is enriched in the primordial germ cells Z2 and Z3 (white arrows) in wild-type and mes-4 mutants but is not detectably concentrated in those cells in mes-2 and mes-6 mutants.
nuclear localization of the other three MES proteins in the germline, suggesting that at all stages MES-4 functions independently from the other MES proteins.
In summary, epistasis analysis suggests that MES-3 associates with MES-2 and MES-6 as a complex in embryos, but the nature of the association is different at different stages of embryogenesis. MES-3 probably does not associate with MES-2 and MES-6 in the germline.
The TSP of MES-3 suggests that MES-3 regulation extends over two generations and is epigenetically heritable: What is the nature of the temperature sensitivity of mes-3(bn21ts)? The bn21 mutation changes a cysteine to a tyrosine. We think the most likely model is that the MES-3 protein produced from the bn21 allele is temperature labile (see Sadler and Novick 1965; Wood et al. 1980). Inactivation induced by high temperature appears to be reversible, since downshifting the embryos can ameliorate the effects of growing their mothers at high temperature.
According to this thermolability model, the results of temperature upshift and downshift b experiments (in which embryos from bn21 mothers placed at 25° as L3 to early L4 larvae were downshifted at different embryonic stages) suggest that functional MES-3 protein must be present during embryogenesis, but not after hatching, for subsequent normal germline development. On the basis of upshift results, the major requirement for MES-3 appears to end between the ~14-cell and ~100-cell stage of embryogenesis, the period when the primordial germ cell P4 is formed but not yet divided into Z2 and Z3. During this period, transcription in P4 is repressed by PIE-1 (Mello et al. 1996; Seydoux et al. 1996). MES-3 protein may function to create or maintain a certain chromatin organization in the primordial germ cell P4, so that when PIE-1 repression is lifted after the division of P4 into Z2 and Z3, the proper pattern of gene expression will be established. This chromatin organization is important for normal germline development and, once established, it apparently is mitotically inherited in the absence of functional MES-3 throughout germline proliferation.
Results from the downshift experiments suggest that the chromatin state induced by MES-3 must be inherited from the previous generation. They further suggest that the cells in which the germline chromatin state must be correctly established in the mother are the germline stem cells. In mes-3(bn21) mothers placed at 25° as L1 to L2 larvae (as done in the downshift a experiment), most germline stem cells would have been formed at 25° and the germline pattern of chromatin would not have been established in them by MES-3. This resulted in almost all their progeny developing into sterile worms, regardless of the temperature at which embryos were cultured. In contrast, when mes-3(bn21) mothers were placed at 25° as L3 to L4 larvae (as done in the downshift b experiment), the stem cell population would have expanded at permissive temperature and would have contained the correct MES-3-induced state of germline chromatin prior to exposure to high temperature. Of the embryos from these mothers, 30% were fertile even when grown at 25° throughout embryogenesis, suggesting that the correct chromatin state can occasionally be propagated through meiosis and mitoses without additional MES-3 action. Additional MES-3 action, provided by downshifting embryos early, led to a majority of embryos developing into fertile adults.
An alternative model is that mes-3(bn21ts) is temperature sensitive for synthesis (TSS; see Sadler and Novick 1965; Wood et al. 1980): Protein produced at permissive temperature is functional at all temperatures, and protein produced at restrictive temperature is nonfunctional at all temperatures. Although we cannot rule out the TSS model, we do not favor it for the following reasons. First, the level of MES-3 is not significantly reduced in mes-3 mothers raised at high temperature from the L1 stage onward. This suggests that the expression and stability of MES-3 are not sensitive to growth temperature. Second, the normal MES-3 distribution is not dependent on MES-2 and MES-6 in the maternal germline and in early embryos, arguing against a requirement for association with MES-2 and MES-6 for MES-3 to be functional. Third, it seems unlikely that the small number of germ cells in an L1–L2 germline would be able to synthesize enough maternal MES-3 to rescue fertility in 30% of their progeny. In fact, the embryo upshift results indicate that a maternal load of MES-3 (produced at permissive temperature) is not sufficient for F1 fertility.
We favor the hypothesis that, for a worm to be fertile, MES-3 must function first in the germline of the worm's mother and then during the early embryonic divisions of the worm. In the maternal (P0) germline, MES-3 maintains or newly establishes a certain chromatin state as the stem cells proliferate. This state is transmitted to the next generation (F1) through meiosis and acts in the next generation as a template. MES-3 in the embryo propagates the maternal chromatin state through the early divisions, ensuring that the primordial germ cells inherit the correct chromatin state for subsequent normal germline development. The fact that the temperature-sensitive period ends during embryogenesis suggests that MES-3's influence is epigenetically inherited through subsequent mitotic divisions. Epigenetic inheritance may be a common feature of chromatin regulation by Polycomb group-related proteins. As discussed in the Introduction, PcG-mediated silencing was found to be mitotically heritable throughout development and to be inherited through meiosis at a certain frequency (Cavalli and Paro 1998). MES-2 and MES-6 are homologs of the PcG proteins E(Z) and ESC (Holdeman et al. 1998; Korf et al. 1998) and are known to form a complex with MES-3. Furthermore, on the basis of analysis of transgene expression (Kelly and Fire 1998), the MES proteins appear to act as transcriptional repressors in the germline, as PcG proteins do in somatic cells. According to our above model, the mechanism of MES regulation may be epigenetically heritable, similar to PcG regulation in Drosophila.
Acknowledgement
We thank Kim House for assistance sequencing mes-3 alleles. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources. This work was supported by NIH grant GM-34059 (to S.S.) and NIH grant GM-51836 (to E.G.).
Footnotes
Communicating editor: T. C. Kaufman
LITERATURE CITED
Author notes
Present address: Center for Cancer Research, Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139.
Present address: Genetics Institute, Cambridge, MA 02140.