-
PDF
- Split View
-
Views
-
Cite
Cite
Satoshi Ansai, Tetsushi Sakuma, Takashi Yamamoto, Hiroyoshi Ariga, Norihito Uemura, Ryosuke Takahashi, Masato Kinoshita, Efficient Targeted Mutagenesis in Medaka Using Custom-Designed Transcription Activator-Like Effector Nucleases, Genetics, Volume 193, Issue 3, 1 March 2013, Pages 739–749, https://doi.org/10.1534/genetics.112.147645
Close - Share Icon Share
Abstract
Transcription activator-like effector nucleases (TALENs) have become powerful tools for targeted genome editing. Here we demonstrate efficient targeted mutagenesis in medaka (Oryzias latipes), which serves as an excellent vertebrate model for genetics and genomics. We designed and constructed a pair of TALENs targeting the medaka DJ-1 gene, a homolog of human DJ-1 (PARK7). These TALENs induced a number of insertions and deletions in the injected embryos with extremely high efficiency. This induction of mutations occurred in a dose-dependent manner. All screened G0 fish injected with the TALENs transmitted the TALEN-induced mutations to the next generation with high efficiency (44–100%). We also confirmed that these TALENs induced site-specific mutations because none of the mutations were found at potential off-target sites. In addition, the DJ-1 protein was lost in DJ-1Δ7/Δ7 fish that carried a TALEN-induced frameshift mutation in both alleles. We also investigated the effect of the N- and C-terminal regions of the transcription activator-like (TAL) effector domain on the gene-disrupting activity of DJ1-TALENs and found that 287 amino acids at the N terminus and 63 amino acids at the C terminus of the TAL domain exhibited the highest disrupting activity in the injected embryos. Our results suggest that TALENs enable us to rapidly and efficiently establish knockout medaka strains. This is the first report of targeted mutagenesis in medaka using TALENs. The TALEN technology will expand the potential of medaka as a model system for genetics and genomics.
SMALL laboratory fish such as medaka (Oryzias latipes) and zebrafish (Danio rerio) are attractive vertebrate models because they are easy to handle and produce large numbers of progeny per generation (Wittbrodt et al. 2002). The medaka is a small freshwater teleost that originally inhabited East Asia and has been used as a model organism since the early 1900s (Aida 1921). There are several advantages to using the medaka in genetics and genomics. First, a high-quality draft genome is available because of its smaller genome (800 Mb), half the size of the zebrafish genome (Kasahara et al. 2007). Second, there are highly polymorphic inbred medaka strains available that can be used for mutagenesis screening and genetic mapping (Naruse et al. 2004). These features significantly contribute to excellent forward genetics studies using medaka, such as the identification of the first sex-determining gene in non-mammalian vertebrates (Matsuda et al. 2002, 2007), a large-scale mutagenesis screen to identify genes acting in diverse developmental processes (Furutani-Seiki et al. 2004), and a fine quantitative trait locus analysis (Kimura et al. 2007).
In addition to these, reverse genetics approaches have been important in demonstrating gene functions and understanding complex biological processes. We previously reported successful targeted mutagenesis in medaka using zinc-finger nuclease (ZFN) technology (Ansai et al. 2012). This system can induce site-specific DNA double-strand breaks that can be repaired by nonhomologous end joining or homology-directed repair, resulting in targeted gene disruptions by insertions and deletions (indels) or in targeted integrations by homologous recombination, respectively (Urnov et al. 2010). Because of its widespread applicability, this approach has been an alternative approach for effective reverse genetics in medaka, in addition to RNA knockdown using antisense morpholino oligonucleotides (Nasevicius and Ekker 2000) and gene knockout by targeting-induced local lesions in genome (TILLING) (Taniguchi et al. 2006). However, there are some limitations to generating a zinc finger domain that binds specifically to a certain genomic target sequence; therefore, the widespread adoption of this technology has been hindered (Bogdanove and Voytas 2011).
Recently, transcription activator-like effector nucleases (TALENs) have been reported as an alternative option for genome editing (Christian et al. 2010; Bogdanove and Voytas 2011; Miller et al. 2011). These enzymes consist of a fusion between a FokI nuclease domain and a transcription activator-like (TAL) effector DNA recognition domain found in plant pathogenic bacteria of the genus Xanthomonas (Christian et al. 2010). The TAL effector domain is composed of 33–35 amino acid repeats, each of which interacts with a single target nucleotide; a polymorphic pair of residues at positions 12 and 13 in each repeat, known as repeat variable di-residues (RVDs), determines its base specificity (Boch et al. 2009; Moscou and Bogdanove 2009). This modularity of DNA recognition allows customization of the TAL effector domain to successfully target the sequence of interest by efficient modular assembly methods (Cermak et al. 2011; Reyon et al. 2012; Sanjana et al. 2012). The TALEN technology has become a powerful approach for genome editing in a number of animal models (Huang et al. 2011; Sander et al. 2011; Tesson et al. 2011; Lei et al. 2012; Liu et al. 2012; Ma et al. 2012; Watanabe et al. 2012).
Here, we demonstrate successful targeted mutagenesis in medaka using TALENs. In this study, we designed and constructed TALENs that target the second exon of the medaka DJ-1 gene, a homolog of the human DJ-1 (PARK7) gene, mutations in which cause autosomal recessive early-onset Parkinson’s disease (Bonifati et al. 2003). Using these TALENs, both somatic and germ-line mutations in the target sequence were specifically induced with high efficiency. We also showed that the DJ-1 protein was lost in homozygous null mutants generated by these TALENs. Moreover, to establish a more effective scaffold of TALENs in medaka, we investigated the effect of the N- and C-terminal regions of the TAL effector domain on the gene-disrupting activity of DJ1-TALENs.
Materials and Methods
Fish
The wild-type medaka used in this study was a d-rR strain (Yamamoto 1953). Fish were handled in accordance with the Animal Research Guidelines at Kyoto University. Fish were maintained in an aquarium with recirculating water in a 14-/10-h day/night cycle at 26°.
Design and construction of TALENs for the DJ-1 gene
The genomic sequence of the DJ-1 locus was identified from the Ensembl medaka genome browser (http://www.ensembl.org/Oryzias_latipes). Potential TALEN target sites in the locus were searched using the TALEN Targeter program (https://tale-nt.cac.cornell.edu/node/add/talen) (Doyle et al. 2012) with the following parameters: (1) spacer length of 14–17, (2) repeat array length of 16–18, and (3) upstream base of T only.
TAL repeats were assembled by the Golden Gate assembly method (Cermak et al. 2011) with slight modifications (Sakuma et al. 2013). Each module containing an RVD was excised from each module plasmid (pNI, pHD, pNN, and pNG vectors) using BsaI-HF (New England Biolabs, Ipswich, MA), and then the fragment was purified using NucleoSpin Gel and PCR Clean-up kit (MACHEREY-NAGEL, Düren, Germany). To generate repeat arrays, the modules were cloned into the dephosphorylated array plasmid (pFUS vectors) using Ligation high Ver.2 (Toyobo, Osaka, Japan). Both the assembled repeat arrays and a last repeat module were excised with Esp3I (Thermo Scientific, Waltham, MA). The fragments were purified using the NucleoSpin Gel and PCR clean-up kit (MACHEREY-NAGEL) and then cloned into expression vectors pCS2TAL3DD or pCS2TAL3RR that contain the SP6 promoter, the truncated N- and C-terminal regions of the TAL effector domain (136 and 63 amino acids, respectively), and either the DD or RR heterodimeric FokI domain (Dahlem et al. 2012).
Construction of truncated TALEN backbone vector
For the addition of a BglII and NheI site at the 3′ end of the SV40 late poly(A) signal in pCS2TAL3DD and pCS2TAL3RR vectors, a fragment containing the polyA signal was amplified by PCR from the pCS2TAL3DD vector using the primers FokI-FW and latepA-RV-MCS (Supporting Information, Table S1) and was cloned into the XbaI/NotI site of the pCSTAL3DD and pCS2TAL3RR vectors.
To generate deletion variants of the TALENs, the TAL effector domain was PCR-amplified from the pTAL3 vector (Cermak et al. 2011). Each N-terminal region of the domain was PCR-amplified using the primer N287FW or N153FW with TAL-R2 (Table S1) and cloned into two Asp718 sites of pCS2TAL3DD and pCS2TAL3RR. The C-terminal region of each domain was amplified using the primer C230RV or C47RV with M13-FW (Table S1) and cloned into the XhoI/BamHI sites of pCS2TAL3DD and pCS2TAL3RR.
RNA preparation and microinjection
The expression vectors for the TALENs were linearized by digestion with either NotI or BglII. Capped RNAs were synthesized using the mMessage mMachine SP6 Kit (Life Technologies, Gaithersburg, MD) and purified using the RNeasy Mini Kit (Qiagen, Valencia, CA). Pairs of RNA for the TALENs were injected into fertilized eggs of the d-rR strain by a microinjection method (Kinoshita et al. 2000).
Somatic mutation analysis in TALEN-injected embryos
Each embryo that was injected with RNA coding for DJ1-TALENs was incubated for 1 hr at 55° in 100 µl lysis buffer containing 150 mM sodium chloride, 0.5% sodium dodecyl sulfate (SDS), 25 mM ethylenediaminetetraacetic acid (EDTA) (pH 8.0), 10 mM Tris–HCl (pH 8.0), and 200 µg/ml of proteinase K. The lysate was extracted with phenol:chloroform and precipitated with ethanol. The DNA precipitate was dissolved in 30 µl deionized water. The genomic region including the target site of the TALENs was amplified using the primers DJ1-FW2 and DJ1-RV1 (Table S1). This reaction contained 2 µl of genomic DNA template, 3 µl of 10× reaction buffer, 0.8 mM of each dNTP, 1.5 mM of MgCl2, 0.2 µM of each primer, and 0.75 unit of HybriPol DNA Polymerase (Bioline, London) in a total volume of 30 µl. The cycling conditions were as follows: one cycle at 95° for 2 min, followed by 30 cycles of 95° for 20 sec, 58° for 30 sec, and 72° for 30 sec. The resulting PCR product was precipitated with ethanol for buffer exchange and was digested at 37° overnight in 10 µl HaeIII digestion solution that contained 1× M buffer and 2.5 unit of HaeIII (Takara Bio, Shiga, Japan). These reactions were analyzed using a microchip electrophoresis system (MCE-202 MultiNA; Shimadzu, Kyoto, Japan) with the DNA-500 Reagent Kit. The molar concentrations of both digested and undigested fragments were quantified using the MultiNA Viewer software.
For sequence analysis in the injected embryos, the DJ-1 genomic region was amplified with TaKaRa Ex Taq (Takara Bio) using the primers DJ1-FW2 and DJ1-RV1 (Table S1). The PCR conditions were as follows: one cycle at 94° for 2 min, followed by 35 cycles of 98° for 10 sec, 56° for 30 sec, and 72° for 30 sec. The PCR amplicons were subcloned into the pGEM-T Easy vector (Promega, Madison, WI) and sequenced using the primer M13-RV (Table S1). The sequencing was performed by Operon Biotechnologies (Tokyo, Japan).
Founder screening
TALEN-injected fish were crossed with wild-type fish of the d-rR strain. Their F1 larvae were lysed individually in 25 µl of alkaline lysis buffer containing 25 mM NaOH and 0.2 mM EDTA (pH 8.0) and incubated at 95° for 10 min. Subsequently, the reaction was neutralized using 25 µl of 40 mM Tris–HCl (pH 8.0). These reactions were centrifuged at 20,000 × g for 3 min, and the supernatants were used as genomic DNA samples. Mutations in the DJ-1 region in each genomic DNA sequence were analyzed by PCR amplification and subsequent HaeIII digestion as described above. Mutant alleles in each mutant F1 fish were determined by direct sequencing of the DJ-1 region that was PCR-amplified using the primers DJ1-FW2 and DJ1-RV1 (Table S1) and purified using the Wizard SV Gel and PCR Clean-Up System (Promega).
Off-target analysis
Potential off-target sites were identified in the medaka genome using the “e-PCR” program (http://www.ncbi.nlm.nih.gov/projects/e-pcr/) as described previously (Lei et al. 2012). In this study, the criteria for determining off-target sites were that up to five non-identical bases and 1-bp gaps can occur in each recognition sequence, and the spacer between two putative recognition regions should be <100 bp.
The genomic region containing the potential off-target site was PCR-amplified from the F1 larvae of TALEN-injected fish using the primers DJ1-off1-FW and DJ1-off1-RV (Table S1). Each amplified product was purified using the Wizard SV Gel and PCR Clean-Up System and directly sequenced using the primer DJ1-off1-FW (Table S1).
Western blot analysis
The head regions from adult medaka were homogenized in cooled phosphate-buffered saline (PBS), pH 7.4, and centrifuged at 20,000 × g for 5 min. The supernatants were boiled in SDS sample buffer containing 50 mM Tris–HCl (pH 6.8), 2% SDS, and 6% 2-mercaptoethanol. Protein samples were separated on a 15% polyacrylamide gel containing SDS and transferred to a polyvinyl difluoride membrane. The membrane was blocked in PBST (PBS with 0.1% Tween-20) containing 1% Block Ace (DS Pharma Biomedical, Suita, Japan) and was probed with rabbit anti-meDJ-1 (Li et al. 2006) diluted 1:1000. The secondary antibody used was anti-rabbit IgG, alkaline phosphatase (AP)-linked antibody (Cell Signaling Technology, Danvers, MA) diluted 1:2000, and signals were developed using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate in AP buffer containing 100 mM Tris–HCl (pH 9.5), 100 mM NaCl, and 50 mM MgCl2.
Statistical analysis
Disruption activities of the truncated TALENs were analyzed with one-way ANOVAs followed by Tukey’s honestly significant difference (HSD) test using the R statistical software (R Development Core Team 2010).
Results
Design and construction of TALENs for the medaka DJ-1 gene
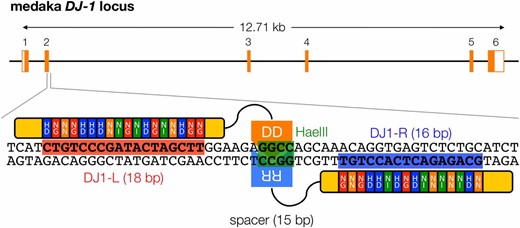
The medaka DJ-1 gene (Ensembl gene no. ENSORLG00000004285), an ortholog of the human DJ-1 (PARK7) gene in medaka, has six exons and encodes 189 amino acids (Li et al. 2006). We scanned potential TALEN target sites in the second exon of the DJ-1 gene using the TALEN Targeter program (Doyle et al. 2012) and identified a target site that consisted of 18 bp of the left binding site (5′-CTGTCCCGATACTAGCTT-3′), 16 bp of the right site (5′-GCAGAGACTCACCTGT-3′), and a 15-bp spacer sequence containing a HaeIII cleavage site (5′-GGAAGAGGCCAGCAA-3′) (Figure 1). This target site also meets all five previously described design guidelines (Cermak et al. 2011). To construct a pair of TALENs cleaving this target site, we assembled RVD modules by the Golden Gate assembly method (Cermak et al. 2011) with slight modifications. All the assembled RVD modules in each site are described in Figure 1.
Genomic structure of the medaka DJ-1 gene (Ensembl gene no. ENSORLG0000004285) and design of DJ1-TALENs. DJ-1 gene has six exons that code 189 amino acids of the DJ-1 protein. DJ1-TALENs were designed to target the second exon of the gene. Red and blue boxes indicate the left and right recognition sequences of the TALENs, respectively. The RVDs of the TAL effector domain for each binding site are shown with the recognition sequences. The large green box in the center of the sequence indicates a cleavage site with HaeIII for mutation analysis.
Efficient mutagenesis with DJ1-TALENs in somatic medaka cells
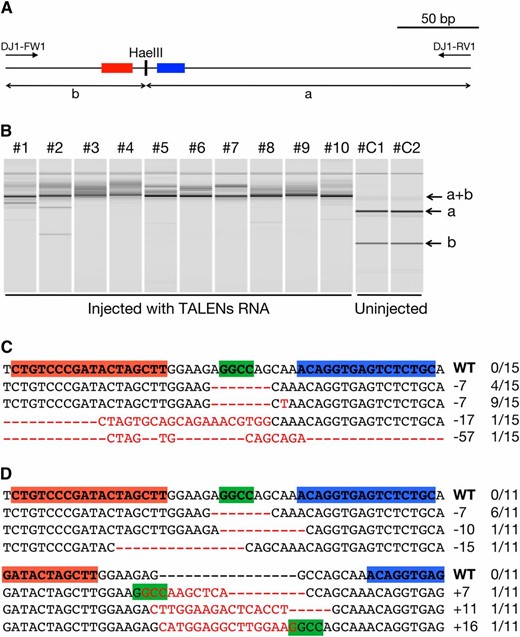
To evaluate whether DJ1-TALENs can induce mutations in the target sequence, 300 ng/µl of RNA coding for the TALENs was injected into fertilized eggs of the d-rR strain. To detect induced mutations, genomic DNA was extracted from each injected embryo at 3 days post fertilization (dpf). The 285-bp long genomic region containing the TALEN target site was PCR-amplified from each genomic DNA, and the amplified product was digested with HaeIII (Figure 2A). Control embryos without the TALENs showed two fragments (199 and 86 bp, a and b, respectively, in Figure 2A) produced by complete digestion with HaeIII. On the contrary, in all 20 TALEN-injected embryos, these two fragments disappeared and one undigested fragment (285 bp, a+b in Figure 2B) distinctly emerged instead, indicating that the HaeIII recognition site in the TALEN target sequence had been mutated. To investigate mutation patterns at the target site, a part of the DJ-1 gene region containing the target site was PCR-amplified from two TALEN-injected embryos (#1 and #2) and was subcloned, and then each clone was sequenced. All 26 clones sequenced had mutated sequences that contained four types of indels (15/15) in embryo #1 (Figure 2C) and six types of indels (11/11) in embryo #2 (Figure 2D), indicating that DJ1-TALENs induced indel mutations around the target site with extremely high efficiency. As shown in Figure 2D, the HaeIII recognition site was newly generated in the mutated allele, and, as a result, two digested fragments of slightly different sizes were observed (Figure 2B, #2).
Somatic mutations in embryos injected with 300 ng/µl RNA for DJ1-TALENs. (A) In the wild type, a 285-bp amplified fragment obtained using primers DJ1-FW2 and DJ1-RV1 is digested into 199-bp (a) and 86-bp (b) fragments by HaeIII. Red or blue boxes indicate the left or right recognition sites of the TALENs, respectively. (B) Gel images of HaeIII-digested fragments analyzed in MultiNA. The TALEN-injected embryos (#1–#10) showed an undigested fragment (a+b), while control embryos that were not injected with TALEN (#C1 and #C2) showed that the intact fragment (a+b) was completely digested into two fragments (a and b). (C and D) Subcloned sequences observed in TALEN-injected embryos #1 and #2, respectively. Red letters or dashes indicate the identified mutations. Red and blue boxes in wild-type (WT) sequences indicate the left and right recognition sites of the TALENs, respectively. Green boxes indicate the HaeIII cleavage site. The sizes of the insertions and deletions are shown to the right of each mutated sequence (−, deletions; +, insertions). Numbers on the right edge indicate the numbers of mutated clones identified from all analyzed clones from each embryo.
DJ1-TALENs can induce mutations in a dose-dependent manner
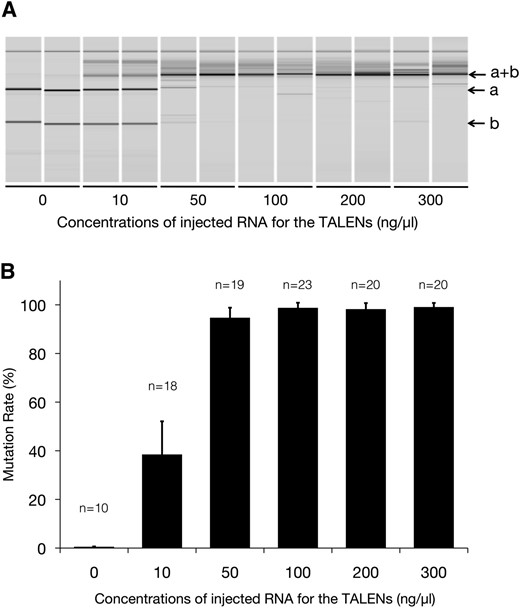
To evaluate the dose dependence of TALEN-mediated gene disruption efficiency, various concentrations (10–300 ng/µl) of RNA coding for the TALENs were injected. Most embryos injected with the TALENs showed no obvious developmental defects at 5 dpf (Table 1). This suggested that TALENs exhibit lower toxicity in the injected medaka embryos than do ZFNs, which induced significant developmental defects or lethality by injection with >10 ng/µl of RNA, although the Sharkey-based DS and RR type of nuclease domains were used in these nucleases (Ansai et al. 2012). Genomic DNA was extracted from each surviving embryo at 5 dpf, and then the HaeIII digestion pattern of the PCR-amplified fragment containing the TALEN target site was analyzed using the MultiNA system. We determined that all TALEN-injected embryos carried mutations at the TALEN-targeted site based on the appearance of the undigested fragment (Figure 3A, a+b). To quantify the disrupting activity at each concentration of TALENs, we calculated the “mutation rate” as the molar concentration of the HaeIII-undigested fragment (3A, a+b) as a percentage of the sum of the molar concentrations of the undigested fragment (Figure 3A, a+b) and the larger digested fragment (Figure 3A, a). As shown in Figure 3B, we found that 10 ng/µl of TALEN RNA induced mutations with low efficiency (38.5% ± 13.6), whereas >50 ng/µl of RNA induced mutations with extremely high efficiency (almost 100%). These results indicate that the DJ1-TALENs dose-dependently induced mutations at the target site.
Survival of embryos injected with various concentrations of RNAs for DJ1-TALENs
| Injected RNA concentration (ng/µl) . | Injected eggs . | Survival at 1 dpf . | Survival at 5 dpf . |
|---|---|---|---|
| 0 | 20 | 20 | 20 |
| 10 | 20 | 19 | 19 |
| 50 | 20 | 20 | 19 |
| 100 | 23 | 23 | 23 |
| 200 | 20 | 20 | 20 |
| 300 | 22 | 20 | 20 |
| Injected RNA concentration (ng/µl) . | Injected eggs . | Survival at 1 dpf . | Survival at 5 dpf . |
|---|---|---|---|
| 0 | 20 | 20 | 20 |
| 10 | 20 | 19 | 19 |
| 50 | 20 | 20 | 19 |
| 100 | 23 | 23 | 23 |
| 200 | 20 | 20 | 20 |
| 300 | 22 | 20 | 20 |
| Injected RNA concentration (ng/µl) . | Injected eggs . | Survival at 1 dpf . | Survival at 5 dpf . |
|---|---|---|---|
| 0 | 20 | 20 | 20 |
| 10 | 20 | 19 | 19 |
| 50 | 20 | 20 | 19 |
| 100 | 23 | 23 | 23 |
| 200 | 20 | 20 | 20 |
| 300 | 22 | 20 | 20 |
| Injected RNA concentration (ng/µl) . | Injected eggs . | Survival at 1 dpf . | Survival at 5 dpf . |
|---|---|---|---|
| 0 | 20 | 20 | 20 |
| 10 | 20 | 19 | 19 |
| 50 | 20 | 20 | 19 |
| 100 | 23 | 23 | 23 |
| 200 | 20 | 20 | 20 |
| 300 | 22 | 20 | 20 |
Dose-dependent mutagenesis by DJ1-TALENs. (A) MultiNA gel images of HaeIII digestion. A PCR fragment containing the TALEN target site was digested with HaeIII. Gel images from two representative embryos injected with 0–300 ng/µl RNA for the TALENs are shown. (B) Mutation rates at each injected TALEN RNA concentration. The mutation rate was calculated as the molar concentration of the undigested fragment (a+b) with HaeIII as a percentage of the sum of molar concentrations of the undigested fragment (a+b) and the larger digested fragment (a). The molar concentration of each fragment was quantified using the MultiNA Viewer software. Columns and error bars represent mean ± SEM. “n” indicates the numbers of embryos analyzed.
Heritable mutations are induced with DJ1-TALENs
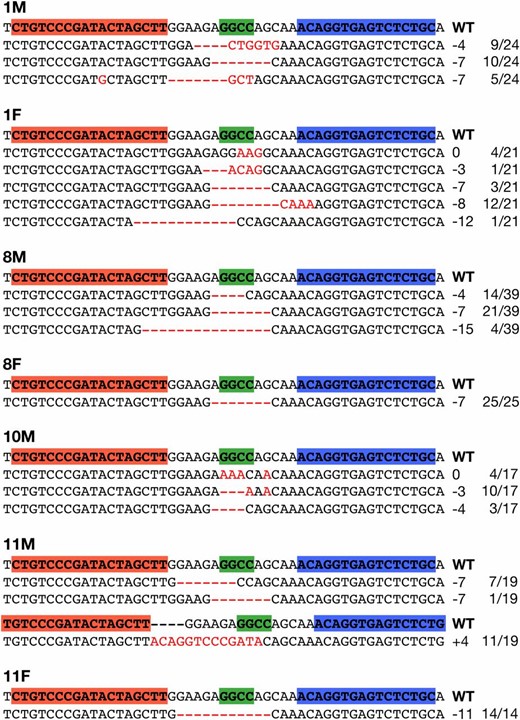
To investigate whether TALEN-mediated mutations are also induced in germ cells and transmitted into subsequent generations, we raised the mutant fish to sexual maturity and analyzed their progeny. Of the 125 eggs injected with 200 ng/µl of TALEN RNA, 95 (76%) hatched normally and 86 (69%) reached adulthood. Seven of the 86 adult G0 fish were crossed with wild-type fish of the d-rR strain, and their F1 progeny were genotyped by PCR amplification and subsequent HaeIII digestion using genomic DNA extracted from each F1 larva. We found that all 7 G0 fish screened transmitted TALEN-induced mutations to their F1 progeny. The germ-line transmission rates of the mutations in each G0 founder ranged from 43.8% (14 of 32; 11F) to 100% (39 of 39; 8M) (Table 2). There was no apparent difference in these rates between the sexes of the G0 founders (Table 2). Subsequently, we performed sequence analysis of all F1 mutant larvae identified by HaeIII genotyping. While 2 of the 7 G0 founders (8 F and 11 F) carried only one type of mutation in their germ cells, the others (1 M, 1 F, 8 M, 10 M, and 11 M) carried from three to five types of mutations in their germ cells (Table 2). All mutations identified from the F1 larvae are shown in Figure 4. These results indicate that DJ1-TALENs induced heritable mutations in injected fish with high efficiency.
Germ-line transmission of TALEN-mediated mutations in each G0 fish
| G0 founder . | . | . | |
|---|---|---|---|
| ID . | Sex . | No. of F1 larvae analyzed . | Mutants (%)a . |
| 1M | Male | 28 | 24 (85.7) |
| 1F | Female | 37 | 21 (56.8) |
| 8M | Male | 39 | 39 (100) |
| 8F | Female | 34 | 25 (73.5) |
| 10M | Male | 28 | 17 (60.7) |
| 11M | Male | 32 | 19 (59.4) |
| 11F | Female | 32 | 14 (43.8) |
| G0 founder . | . | . | |
|---|---|---|---|
| ID . | Sex . | No. of F1 larvae analyzed . | Mutants (%)a . |
| 1M | Male | 28 | 24 (85.7) |
| 1F | Female | 37 | 21 (56.8) |
| 8M | Male | 39 | 39 (100) |
| 8F | Female | 34 | 25 (73.5) |
| 10M | Male | 28 | 17 (60.7) |
| 11M | Male | 32 | 19 (59.4) |
| 11F | Female | 32 | 14 (43.8) |
The values shown are the numbers of mutant F1 larvae as determined by PCR amplification and subsequent HaeIII digestion.
| G0 founder . | . | . | |
|---|---|---|---|
| ID . | Sex . | No. of F1 larvae analyzed . | Mutants (%)a . |
| 1M | Male | 28 | 24 (85.7) |
| 1F | Female | 37 | 21 (56.8) |
| 8M | Male | 39 | 39 (100) |
| 8F | Female | 34 | 25 (73.5) |
| 10M | Male | 28 | 17 (60.7) |
| 11M | Male | 32 | 19 (59.4) |
| 11F | Female | 32 | 14 (43.8) |
| G0 founder . | . | . | |
|---|---|---|---|
| ID . | Sex . | No. of F1 larvae analyzed . | Mutants (%)a . |
| 1M | Male | 28 | 24 (85.7) |
| 1F | Female | 37 | 21 (56.8) |
| 8M | Male | 39 | 39 (100) |
| 8F | Female | 34 | 25 (73.5) |
| 10M | Male | 28 | 17 (60.7) |
| 11M | Male | 32 | 19 (59.4) |
| 11F | Female | 32 | 14 (43.8) |
The values shown are the numbers of mutant F1 larvae as determined by PCR amplification and subsequent HaeIII digestion.
TALEN-induced mutations observed in F1 larvae from seven G0 founders. Red letters or dashes indicate the identified mutations. Red and blue boxes in the wild-type (WT) sequences indicate the left and right recognition sites of the TALENs, respectively. Green boxes indicate the HaeIII cleavage site. The sizes of the insertions and deletions are shown to the right of each mutated sequence (−, deletions; +, insertions). The numbers on the right edge indicate the numbers of larvae carrying each mutated sequence among all sequenced larvae.
Specificity of mutations induced by DJ1-TALENs
For accurate genome editing, it is important to induce mutations site-specifically with TALENs. We searched for potential off-target sites of DJ1-TALENs using the e-PCR program and identified 13 candidates for potential off-target sites in the medaka genome (Figure S1). It has been reported that the scaffold of our TALENs had less disrupting activity when the spacers between two binding sites were >24 bp long (Miller et al. 2011). Since 12 of 13 candidates have >100 bp in their spacers (Figure S1), it is unlikely that the TALENs induce mutations at these sites. We analyzed one candidate site (chromosome 9: 15,931,407–15,931,476) that had 10-bp mismatches and 2-bp gaps in the recognition sequences and the shortest 32-bp spacer among the 13 candidates. The genomic region containing the off-target candidate site was PCR-amplified from 12 F1 larvae of four founders (1 M, 1 F, 8 M, and 8 F) that carried independent mutations in their DJ-1 regions (Figure 4), and then the amplified product was subjected to direct sequencing. As a result, we identified no mutation at the site, indicating that DJ1-TALENs have no potential off-target site with significant similarity to their target site in medaka genome.
Homozygous DJ-1 mutants generated with DJ1-TALENs lacked DJ-1 protein
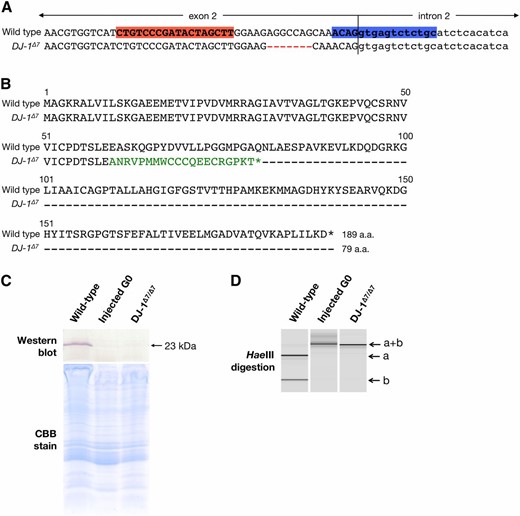
Next, we investigated whether genomic mutations with DJ1-TALENs affect the production of the DJ-1 protein. Interestingly, we found that a frameshift mutation with the same 7-bp deletions (5′-AGGCCAG-3′ in the spacer region) was identified in F1 larvae from five G0 founders (Figure 4; Figure 5A). This frameshifted mutant allele (named DJ-1Δ7) yields a truncated DJ-1 protein due to an additional region of altered translation (Figure 5B). We therefore generated null mutants that carried DJ-1Δ7 in both alleles by crossing the founders 8M and 8F. To analyze the production of the DJ-1 protein, extracts from the head region of this F1 null mutant, a G0 fish injected with 200 ng/µl TALEN RNA and a wild-type fish of the d-rR strain were subjected to Western blot analysis with the anti-medaka DJ-1 antibody produced previously (Li et al. 2006). An immunoreactive band with a molecular mass of 23 kDa was detected in the wild-type fish, while no signals were detected in the F1DJ-1Δ7/Δ7 fish (Figure 5C). This result indicated that loss-of-function mutations were induced by the TALENs. It is noteworthy that even the G0 fish injected with the TALENs completely lacked DJ-1 protein (Figure 5D). It is reasonable to assume that, in the G0 fish, the DJ-1 gene was completely disrupted by TALEN injection (Figure 3).
Expression of the DJ-1 protein in homozygous mutant fish. (A) Nucleotide sequences from the wild-type and mutant DJ-1Δ7 alleles. Red dashes indicate the TALEN-induced deletion. Red and blue boxes indicate the left and right recognition sites of DJ1-TALENs, respectively. Uppercase and lowercase letters in the sequences indicate the second exon and second intron sequences of the DJ-1 gene, respectively. (B) Alignment of the amino acid sequences of wild-type (accession no. AB193829) and truncated medaka DJ-1 proteins led by the DJ-1Δ7 frameshifted mutation. The DJ-1Δ7 product has an additional region of altered translation, which is indicated by green letters. (C) Western blot analysis using anti-medaka DJ-1 antibody. Homogenates prepared using the head regions of each fish were used (see Materials and Methods for details). The predicted signal (23 kDa) was detected in the wild-type lysate. No signals were detected in both injected G0 and DJ-1Δ7/Δ7 lysates, although the Coomassie Brilliant Blue (CBB)-stained gel showed evenly extracted protein bands among the three samples (bottom). (D) MultiNA gel images of HaeIII digestion. Genomic DNA was extracted from the posterior part of each fish whose head region was analyzed with Western blot in C. PCR fragments containing the target site of DJ1-TALENs were digested with HaeIII. In the wild type, the intact band (a+b) was completely digested into two fragments (a and b), whereas both the injected G0 fish and the DJ-1Δ7/Δ7 mutant exhibited an undigested fragment (a+b). (C and D) “Wild-type”: without TALEN-injection; “injected G0 fish”: injected with 200 ng/µl RNA for the TALENs; “DJ-1Δ7/Δ7”: F1 fish harboring a 7-bp deletion (DJ-1Δ7) in both alleles.
Disrupting activity of DJ1-TALENs largely depends on the length of the N- and C-terminal regions of the TAL effector domain
The length of fully assembled TAL effectors (>800 amino acids) makes it difficult to handle their DNA constructs and RNA transcripts. Therefore, there have been several efforts to identify the minimal N- and C-terminal regions of the TAL effector domain for efficient DNA cleavage (Miller et al. 2011; Mussolino et al. 2011). In this study, we used the pCS2TAL3DD and pCSTAL3RR vectors for TALEN RNA synthesis, both of which harbor 136 or 63 amino acids in the N- or C-terminal regions of the TAL effector domain. The TALENs generated from these vectors induced site-specific mutation with high efficiency not only in the medaka (in this study) but also in the zebrafish (Dahlem et al. 2012).
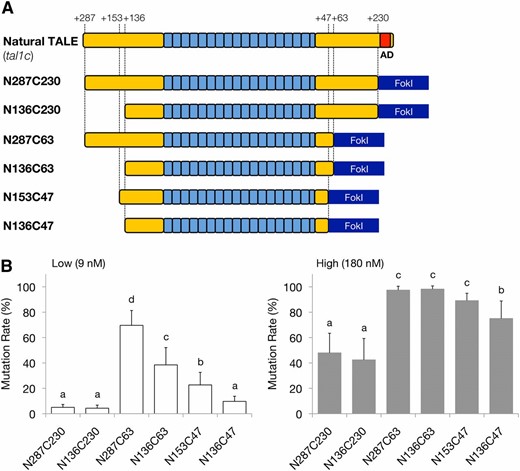
To identify more efficient scaffolds of TALENs for in vivo genome cleavage, six types of deletion variants of DJ1-TALENs were constructed (Figure 6A). N287 and C230 scaffolds were comparable to the original scaffold of TALENs (Christian et al. 2010; Cermak et al. 2011), while N153 and C47 were comparable to one of the effective deletion variants (named “NC” type) (Mussolino et al. 2011). Each pair of RNA sequences for these TALENs was injected into fertilized eggs and did not show significant toxicity to the embryo (summarized in Table 3). From each 5-dpf embryo, the genomic DNA fragment containing the target site of the DJ1-TALENs was PCR-amplified and was subjected to subsequent HaeIII digestion to assess the “mutation rate” as described previously (Figure 6B). As shown in Figure 6B, with higher concentrations of TALEN-RNA (180 nM), N287C230 and N136C230 induced mutations in about 50% of the injected embryos. On the other hand, other constructs (N287C63, N136C63, N153C47, and N136C47) induced higher mutation rates (over 75%). This trend was more remarkable in the case of lower amounts of TALEN-RNA (9 nM). Among the N136 scaffolds (N136C230, N136C63, and N136C47 in Figure 6A), we found that 63 amino acids of the C-terminal region of the domain showed the highest disrupting activity (38.5 ± 13.6%). In addition, among the C63 scaffolds, N287C63 showed a higher disruption rate (69.6 ± 11.7%) than did N136C63 (38.5 ± 13.6%). This tendency was also observed in the C47 scaffolds: N153C47 showed a higher disruption rate (22.7 ± 9.9%) than did N136C47 (9.7 ± 4.1%). As above, in our experiments, the longer N-terminal regions showed more disruption activity.
Effects of the N and C termini of the TAL effector domain on the gene-disrupting activities of deletion variants of DJ1-TALENs. (A) Schematic of deletion variants in the TAL effector domain. A natural TALE (tal1c) has 287 and 278 amino acids in the N- and C-terminal regions of the TAL effector domain, respectively. The transcriptional activation domain (AD) in the C-terminal region is highlighted in red. The names of the deletion variants generated by PCR amplification are indicated on the left side. The numbers of remaining amino acids of the N- and C-terminal regions relative to the position of the repeat modules are shown above. (B) Disruption activities of the truncated TALENs, shown as the mutation rate in the injected embryos. (Left) Mutation rates in the embryos injected with “low” concentrations of RNAs (9 nM) containing a mole ratio equal to 10 ng/µl of RNA for the N136C63 scaffold of the TALENs. (Right) Mutation rates in the embryos injected with “high” concentrations of RNAs (180 nM) containing a mole ratio equal to 200 ng/µl of the N136C63 scaffold. Mutation rate was calculated as the molar concentration of the undigested fragment with HaeIII as a percentage of the sum of the molar concentrations of the undigested fragment and the larger digested fragment. The molar concentration of each fragment was quantified using the MultiNA Viewer software. Columns and error bars represent mean ± SEM (n = 12). The differnt letters at the top of the columns indicate significant differences (P < 0.05; one-way ANOVA and Tukey’s HSD test).
Results of microinjection with the deletion series of DJ1-TALENs RNA
| Scaffold . | RNA concentration (nM) . | Injected eggs . | Survival at 1 dpf . |
|---|---|---|---|
| N287C230 | Low (9) | 24 | 24 |
| High (180) | 24 | 16 | |
| N136C230 | Low (9) | 23 | 23 |
| High (180) | 24 | 24 | |
| N287C63 | Low (9) | 24 | 23 |
| High (180) | 24 | 20 | |
| N136C63a | Low (9) | 20 | 19 |
| High (180) | 20 | 20 | |
| N153C47 | Low (9) | 24 | 21 |
| High (180) | 24 | 22 | |
| N136C47 | Low (9) | 24 | 22 |
| High (180) | 22 | 19 |
| Scaffold . | RNA concentration (nM) . | Injected eggs . | Survival at 1 dpf . |
|---|---|---|---|
| N287C230 | Low (9) | 24 | 24 |
| High (180) | 24 | 16 | |
| N136C230 | Low (9) | 23 | 23 |
| High (180) | 24 | 24 | |
| N287C63 | Low (9) | 24 | 23 |
| High (180) | 24 | 20 | |
| N136C63a | Low (9) | 20 | 19 |
| High (180) | 20 | 20 | |
| N153C47 | Low (9) | 24 | 21 |
| High (180) | 24 | 22 | |
| N136C47 | Low (9) | 24 | 22 |
| High (180) | 22 | 19 |
The results from the N136C63 scaffold are the same as shown for 10 and 200 ng/µl in Table 1.
| Scaffold . | RNA concentration (nM) . | Injected eggs . | Survival at 1 dpf . |
|---|---|---|---|
| N287C230 | Low (9) | 24 | 24 |
| High (180) | 24 | 16 | |
| N136C230 | Low (9) | 23 | 23 |
| High (180) | 24 | 24 | |
| N287C63 | Low (9) | 24 | 23 |
| High (180) | 24 | 20 | |
| N136C63a | Low (9) | 20 | 19 |
| High (180) | 20 | 20 | |
| N153C47 | Low (9) | 24 | 21 |
| High (180) | 24 | 22 | |
| N136C47 | Low (9) | 24 | 22 |
| High (180) | 22 | 19 |
| Scaffold . | RNA concentration (nM) . | Injected eggs . | Survival at 1 dpf . |
|---|---|---|---|
| N287C230 | Low (9) | 24 | 24 |
| High (180) | 24 | 16 | |
| N136C230 | Low (9) | 23 | 23 |
| High (180) | 24 | 24 | |
| N287C63 | Low (9) | 24 | 23 |
| High (180) | 24 | 20 | |
| N136C63a | Low (9) | 20 | 19 |
| High (180) | 20 | 20 | |
| N153C47 | Low (9) | 24 | 21 |
| High (180) | 24 | 22 | |
| N136C47 | Low (9) | 24 | 22 |
| High (180) | 22 | 19 |
The results from the N136C63 scaffold are the same as shown for 10 and 200 ng/µl in Table 1.
Discussion
We demonstrated successful targeted mutagenesis in medaka using TALENs: that is, TALENs targeted to the medaka DJ-1 gene could induce mutations in both the somatic and the germ cells of the injected fish with extremely high efficiency. In addition, we also showed that the DJ-1 protein was lost in null mutant fish that carried the TALEN-induced frameshifted mutation in both alleles. These results revealed that the TALEN technology is an available and effective tool for reverse genetics in medaka. This is the first report to describe targeted disruption of an endogenous gene in medaka using engineered TALENs.
TALENs can simply and efficiently produce gene knockout strains with RNA introduction by conventional microinjection methods. In this study, all the screened G0 founders that were injected with the TALENs (7/7; 100%) carried mutations in their germ cells, and these mutation events were induced in their germ cells with high frequency (68.6% on average; Table 2). This frequency was equal or superior to those in our mutagenesis study using ZFNs in medaka (46.9% on average) (Ansai et al. 2012) and in previous studies using TALENs in zebrafish (8.3–66.0% on average) (Bedell et al. 2012; Cade et al. 2012; Dahlem et al. 2012).
Detailed investigation of the mutations in F1 larvae revealed that up to five types of mutations were induced in the germ cells of each TALEN-injected G0 founder (Figure 4). Taking the high mutation rate mentioned above into consideration, these facts indicate that mutation events occurred independently in a number of embryonic cells in early embryogenesis. From a practical standpoint, this suggests that a number of mutant strains can be obtained from each founder; therefore, determination of mutant alleles by sequencing in each F1 fish is required before mating the F1 fish for the establishment of mutant strains.
Highly efficient induction of mutations by the TALENs also occurred in the somatic cells of injected G0 fish. Interestingly, in a G0 fish injected with 200 ng/µl TALEN RNA, the DJ-1 protein was reduced to undetected levels in Western blot analysis (Figure 5C) in accordance with the fact that lesions were induced at the target site by the TALENs in the G0 fish (Figure 5D). Mosaic patterns of mutations were observed in the RNA-injected fish, as in our previous study using ZFNs (Ansai et al. 2012); however, in this study, both mutation analysis with HaeIII digestion and sequence analysis suggested that the TALENs induced the complete disruption of the targeted gene in the injected fish (G0) (Figures 2 and 3) as well as the complete loss of the protein (Figure 5D). These results strongly suggest that highly effective TALENs can induce complete gene knockout in the TALEN-injected generation as previously reported in zebrafish (Bedell et al. 2012). This will make it possible to perform gene knockout experiments without mating or the establishment of mutant strains, shortening the duration of experiments. In addition, TALEN-mediated gene lesions and the induced protein losses in the injected fish produced by them are retained throughout their life. This is an additional advantage of using TALEN-injected fish, compared with gene knockdown experiments using antisense morpholinos that interfere with gene translation only during the early developmental stages.
On the other hand, when we disrupt genes whose functions are essential for embryonic development or reproduction, such highly efficient induction of mutations that cause complete loss of the targeted gene products is likely to lead to embryonic lethality or sterility in the injected fish. Hence, it may happen that highly effective TALENs make it difficult to obtain heterozygous mutants that exhibit no or weaker defects. However, in this study, we showed that the gene-disrupting activity of DJ1-TALENs was significantly reduced by injecting a low concentration (10 ng/µl) of RNA (39%), whereas >50 ng/µl of RNA efficiently induced mutations (almost 100%) (Figure 3). This result suggests that dilution of the injected RNA enables us to avoid the induction of bi-allelic mutations and thereby lethality or sterility in the injected fish; therefore, this approach will make it possible to establish gene knockout strains with disruptions in genes with essential functions.
When establishing null mutant strains using random mutagenesis approaches such as the TILLING method, time-consuming and laborious procedures are required to remove unwanted nonspecific mutations, for example, repeated backcrossing, which takes ∼12 or more months (Kamei 2009). Meanwhile, because the DNA-binding domain in each TALEN has a sufficiently long binding sequence (recognizing from 16 to 18 bp) that exhibits high DNA-binding specificity, it is likely that there are few off-target effects caused by mutagenesis of potential off-target sites of the TALENs. In fact, we could identify no candidate for an off-target site for DJ1-TALENs using the e-PCR program with the criteria described previously (Lei et al. 2012). Upon analyzing a potential off-target site identified with less stringent criteria (allowing up to a 5-bp mismatch and a 1-bp gap in each recognition site) (Figure S1), no mutations were found at the site in mutated F1 larvae. These results suggest that TALENs induce mutations site-specifically in medaka. Because of this high specificity, backcrossing to reduce off-target defects is not necessarily required when establishing mutant strains using TALENs. In addition to frequently inducing the same mutation, such as DJ-1Δ7 (Figure 5A), this suggests that homozygous null mutants (e.g., DJ1Δ7/Δ7 in this study) can be generated in the F1 generation by crossing between the TALEN-injected G0 founders (Figure 5). This is one of the advantageous characteristics of targeted mutagenesis with the TALEN system: we can obtain and begin to analyze homozygous mutants in the short time period of only 3–4 months.
Despite the high specificity of the TALENs, it was reported that a closely related paralog of the targeted gene was disrupted by TALENs in the zebrafish (Dahlem et al. 2012). As well as the zebrafish, the medaka also has >2000 pairs of paralogs derived from whole-genome duplication in teleosts (Kasahara et al. 2007). This is likely to make it difficult to disrupt and analyze the paralogs independently. Therefore, it is important to search the paralogs of the targeted genes that may carry very similar sequences to the target sequence of the designed TALENs for medaka endogenous genes.
Although Western blot analysis with the anti-medaka DJ-1 antibody indicates that the DJ-1 protein was lacking in both the injected G0 fish and the F1DJ-1Δ7/Δ7 fish (Figure 5C), we did not observe developmental or behavioral defects (data not shown). It was reported that in DJ-1-deficient mice and zebrafish, loss of dopaminergic (DA) neurons is intensely induced by neurotoxins and oxidative stress (Kim et al. 2005; Bretaud et al. 2007). These observations suggest that a more detailed investigation of the relationship between the loss of DA neuron and other factors such as external stresses and genetic background is required to explain the functions of DJ-1 in medaka.
In this study, we investigated the effect of the N- and C-terminal regions of the TAL effector domain on the gene-disrupting activity of DJ1-TALENs and found that 287 amino acids of the N-terminal region (N287) and 63 amino acids of the C-terminal region (C63) showed the highest activities among six types of deletion variants of the TALENs (Figure 6). It is likely that the C63 scaffolds efficiently matched the 15-bp spacer sequences used in this study because previous reports suggested that the C-terminal region determines the length of spacer sequences of the TALENs (Miller et al. 2011; Mussolino et al. 2011). On the other hand, it remains difficult to explain why both the N287 and the N153 scaffolds had higher activities than the N136 scaffold (Figure 6B) because most previous TALEN studies adopted only the N136 scaffold, in which the deletion of the first 152 amino acids was considered to specifically contribute to transport into plant cells (Szurek et al. 2002). It is likely that the first 152 amino acids play some role in the DNA cleavage activity of TALENs, such as facilitation of DNA binding and/or stabilization of the proteins. More detailed investigation focused on the functions of the N-terminal region is required to explain our results.
In the medaka, there are a number of inbred strains that show diverse morphological, behavioral, and genetic characteristics (Naruse et al. 2004). The combination of TALEN-mediated genome editing and these strains make it possible to fully examine the relationship between the targeted gene and the genetic background. Moreover, TALENs also make it possible to disrupt specific functional motifs or domains and induce targeted genome modifications with homology-directed repair. These applications of TALEN technology will further expand the potential of the medaka fish as an excellent model system for genetics.
Acknowledgments
We thank Kazuyuki Hoshijima and David Grunwald for providing the pCS2TAL3DD and pCS2TAL3RR vectors. We also thank Daniel F. Voytas and Addgene for providing the Golden Gate TALEN and TAL Effector Kit (#1000000016).
Literature Cited
Kamei, Y., 2009 Tilling (gene knockout), pp. 373–381 in Medaka: Biology, Management, and Experiment Protocols, edited by M. Kinoshita, K. Murata, K. Naruse, and M. Tanaka. Wiley-Blackwell, Ames, IA.
Footnotes
Communicating editor: D. Voytas
Author notes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.112.147645/-/DC1.