-
PDF
- Split View
-
Views
-
Cite
Cite
Margot T Goldberg, Rachel B Spigler, Tia-Lynn Ashman, Comparative Genetic Mapping Points to Different Sex Chromosomes in Sibling Species of Wild Strawberry (Fragaria), Genetics, Volume 186, Issue 4, 1 December 2010, Pages 1425–1433, https://doi.org/10.1534/genetics.110.122911
Close - Share Icon Share
Abstract
Separate sexes have evolved repeatedly from hermaphroditic ancestors in flowering plants, and thus select taxa can provide unparalleled insight into the evolutionary dynamics of sex chromosomes that are thought to be shared by plants and animals alike. Here we ask whether two octoploid sibling species of wild strawberry—one almost exclusively dioecious (males and females), Fragaria chiloensis, and one subdioecious (males, females, and hermaphrodites), F. virginiana—share the same sex-determining chromosome. We created a genetic map of the sex chromosome and its homeologs in F. chiloensis and assessed macrosynteny between it and published maps of the proto-sex chromosome of F. virginiana and the homeologous autosome of hermaphroditic diploid species. Segregation of male and female function in our F. chiloensis mapping population confirmed that linkage and dominance relations are similar to those in F. virginiana. However, identification of the molecular markers most tightly linked to the sex-determining locus in the two octoploid species shows that, in both, this region maps to homeologues of chromosome 6 in diploid congeners, but is located at opposite ends of their respective chromosomes.
SEX chromosomes have evolved multiple times in diverse taxa of both plants and animals (Fraser and Heitman 2005), and theory predicts a stepwise transition from autosomes in hermaphroditic organisms to sex chromosomes in organisms with separate sexes (Ohno 1967; Bull 1983). In the first step, a male-sterility mutation gives rise to females, followed by one or more female sterility mutations, which are favored when they increase male function and are closely linked to the initial male sterility mutation (Charlesworth and Charlesworth 1978). Later, selection for recombination suppression between these loci to reduce the production of neuters as well as the gradual degeneration of the nonrecombining chromosome eventually lead to heteromorphism (reviewed in Charlesworth 2008). Detailed studies in a number of species have provided convincing evidence for these initial steps, including linked sterility loci, recombination suppression, and its genomic consequences (e.g., Westergaard 1958; Bachtrog 2005; Telgmann-Rauberet al. 2007; Filatov 2008; Unoet al. 2008a; Yuet al. 2008b; Spigleret al. 2010), whereas comparative studies have exposed additional complexity in the evolutionary dynamics of sex-determining chromosomes. For example, sex chromosomes not only have arisen de novo from different autosomes and with different dominance relations (i.e., XY or ZW) in related species (e.g., fish: Cnaaniet al. 2008; Rosset al. 2009; frogs: Ogataet al. 2007; Unoet al. 2008b), but also well-established sex chromosomes are subject to change, e.g., the fusion of an ancestral sex chromosome to an autosome leading to the formation of neo-sex chromosomes in Drosophila miranda (White 1973; Steinemann and Steinemann 1998; Bachtrog and Charlesworth 2000).
While many studies have involved animals, flowering plants are an excellent platform for studying sex chromosome evolution. Not only have separate sexes (dioecy) arisen repeatedly from hermaphroditism in angiosperms (Charlesworth 1985, 2002; Renner and Ricklefs 1995), but also, when present, sex chromosomes in angiosperms are often young (e.g., <15 MYA: Bergeroet al. 2007; Yuet al. 2008b) and may not have undergone the later steps of sex chromosome evolution. Furthermore, comparative work in plants has uncovered sex chromosomes that are as diverse and dynamic as those in animals. For example, phylogenetic work has shown that dioecy has evolved independently twice in Silene (Desfeuxet al. 1996; Mrackovaet al. 2008). Furthermore, sex chromosomes have diverged within one section of the genus, with species Silene diclinis possessing neo-sex chromosomes, while two sister species (S. latifolia and S. dioica) have only the ancestral state (Howellet al. 2009). Despite the wide phylogenetic distribution of dioecy and sex chromosomes in flowering plants (Charlesworth 2002), only a handful have been the subject of genetic mapping studies (grape: Margueritet al. 2009; asparagus, Reamon-Buettneret al. 1998; Telgmann-Rauberet al. 2007; papaya: Sonduret al. 1996; Maet al. 2004; Chenet al. 2007; kiwifruit: Testolinet al. 2001), and even fewer have been the subject of comparative analyses that can inform on other evolutionary dynamics (e.g., Rumex: Navajas-Perezet al. 2005; Cunadoet al. 2007; Silene: Howellet al. 2009; papaya: Yuet al. 2008a; Wuet al. 2010).
Wild strawberries (Fragaria, Rosaceae) show the entire range of sexual systems—from hermaphroditism to dioecy (Staudt 1989); diploids are almost exclusively hermaphroditic (e.g., Fragaria vesca and Fragaria nubicola, but gynodioecy exists in one subspecies of F. vesca), whereas polyploids are predominantly sexually polymorphic (for example, both dioecious Fragaria chiloensis and subdioecious Fragaria virginiana are octoploids: Hancock and Bringhurst 1979; Staudt 1989; Ashman 1999). Recent genetic maps have revealed macrosynteny between the diploid and octoploid genomes (Rousseau-Gueutinet al. 2008; Sargentet al. 2009; Spigleret al. 2010), allowing the autosomal homeolog of the proto-sex chromosome of subdioecious F. virginiana to be identified (Spigleret al. 2010). Our study takes advantage of this published work to begin to answer the overarching question: Is the sex-determining region of dioecious F. chiloensis syntenous with that of its sibling species, subdioecious F. virginiana?
F. chiloensis and F. virginiana are allo-allopolyploids (Bringhurst 1990) with disomic inheritance (2n = 8x = 56) (Ashleyet al. 2003; Lerceteau-Köhleret al. 2003; Rousseau-Gueutinet al. 2008). The two species share a very recent common ancestor (Rousseau-Gueutinet al. 2009; Njuguna 2010) and are thought to have diverged only after migration across the Bering Strait to northwestern North America from Asia (Staudt 1999), where F. virginiana dispersed eastward and F. chiloensis southward. While F. virginiana populations have varying frequencies of females, hermaphrodites, and males (Ashman 1999), F. chiloensis is primarily dioecious, although fruit-producing male-fertile individuals (i.e., hermaphrodites) have been reported in some populations (Hancock and Bringhurst 1979; Staudt 1999). The latter commonly produce only a single fruit from many flowers and do so early in the season. In contrast, the high-fruiting hermaphrodites that are known to exist in F. virginiana (Ashman 1999; Bishopet al. 2010) are extremely rare in F. chiloensis (Hancock and Bringhurst 1979). It has been assumed that both species share the same genetic sex-determination system, i.e., a single locus or gene region with three alleles (femaleness “F” dominant to hermaphroditism “h” and maleness “m”: Ahmadi and Bringhurst 1991). However, this model was re-evaluated for F. virginiana by scoring male and female function separately and using genetic mapping (Spigleret al. 2008). This work revealed that two linked but recombining sex function loci, each with a sterility allele (for male function, sterility “A” is dominant to fertility “a”; for female function, sterility “g” is recessive to fertility “G”), determine sexual function in this species (Spigleret al. 2008). Here we use a similar mapping approach to test whether the sex-determining region of F. chiloensis is syntenous with that of F. virginiana and whether they might share a two-locus system. Given the close phylogenetic relationship of the two species (Rousseau-Gueutinet al. 2009; Njuguna 2010), we expected that the sex-determining loci in F. chiloensis would prove to be orthologous to those in F. virginiana and that the main difference is that F. chiloensis has increased recombination suppression between the genes involved (Spigleret al. 2010), i.e., that these two species represent two stages of the step-wise evolution of a sex chromosome proposed by Charlesworth and Charlesworth (1978). An alternative is that a sex-determining region either arose de novo in separate autosomes or moved to another chromosome in one of the species as the result of rearrangement. The probability of either mechanism may be heightened in polyploid species if genome doubling and merger increases the probability of rearrangements or mutations.
With this study, we sought to answer the following two questions: First, are the linkage and dominance relations of the F. chiloensis sex-determining system the same as those of F. virginiana? Second, is the sex-determining chromosome of F. chiloensis in the same homeologous group as it is in F. virginiana, and if so, is the sex-determining region of F. chiloensis at the same chromosomal location as it is in F. virginiana?
MATERIALS AND METHODS
Creation and cultivation of a F. chiloensis mapping population:
The maternal parent (GP33) used to create the mapping population was a female F. chiloensis subsp. lucida, originally collected from Honeyman Memorial State Park, Florence, Oregon (43.93166667N, 124.11083333W) that we obtained from the National Clonal Germplasm Repository (accession no. PI 612489). The paternal parent (SAL3) was a male-fertile F. chiloensis subsp. pacifica that was collected from Salishan, Oregon (44.9166869N, 124.0270281W), and had low fruit set in the greenhouse (10% of flowers yielded a fruit). Thus, the paternal parent represents one of the rare instances of a low-fruit-producing hermaphrodite in this primarily dioecious species (Hancock and Bringhurst 1979). This cross ostensibly represents one between the two North American subspecies of F. chiloensis. However, because they are distinguished by just one character [angle of pubescence: Catling and Porebski 1998; Staudt 1999)] and share all other morphometric characters (Hancocket al. 2004), molecular genetic diversity (Hokansonet al. 2006), and ranges (Staudt 1999), some investigators (Hokansonet al. 2006) have proposed that the two are one subspecies.
We pollinated flowers of GP33 by hand with pollen from SAL3 in April 2009 and planted 137 of the resultant seeds in June 2009. We transplanted seedlings into 3-inch-square pots filled with a 2:1 mixture of Fafard #4 (Conrad Fafard) and sand. During the course of the study, the plants received a total of 513 mg of granular Nutricote 13:13:13 N:P:K fertilizer (Chisso-Asahi Fertilizer) and were protected from pests as needed. In addition, we produced two clones of each of the parents in November 2009. All plants were grown under cool temperatures (12°/18° night/day) and 10- to 12-hr days throughout the majority of the flowering period. This study includes a random selection of 92 progeny that were scored for genetic markers as described below.
Sex-expression assessment:
We assessed male function on at least two flowers per plant two times during flowering. Plants with large, bright-yellow anthers that visibly released pollen were considered “male fertile,” while plants with vestigial white or small, pale-yellow anthers that neither dehisced nor showed mature pollen were considered “male sterile.” Subsamples of all types of anthers as well as any others that were in question were examined with the aid of a compound microscope for the presence of dehiscence and well-formed pollen. To ensure full potential seed and fruit set, we hand-pollinated all plants three times per week with outcrossed pollen. We estimated female function for each individual as the proportion of flowers that produced fruit (“fruit set”) by dividing the total number of fruits by the total number of flowers produced. To be consistent with previous qualitative mapping of sex in F. virginiana (Spigleret al. 2008), we considered plants with ≥5% fruit set as “female fertile” and those with <5% as “female sterile.” We scored both sex functions on all plants that flowered except for two plants that lacked fruit set data.
DNA extraction, PCR, and marker analysis:
We extracted DNA from young leaf tissue from two replicates of the parents and one from each progeny using the Qiagen DNeasy 96 Plant Kit (Qiagen). DNA was diluted to 0.03 ng/ml with distilled deionized water for PCR reactions. Because we were interested in testing synteny, rather than creating a genetic map of the whole genome, we used 56 primer pairs for DNA markers (SSRs or genes) that have mapped either to the homeologous group (HG) in F. virginiana to which the proto-sex chromosome belongs (i.e., HG VI; Spigleret al. 2010) or to the homeologous linkage group (LG) in a diploid interspecific cross (i.e., LG 6; Sargentet al. 2006, 2008). PCR was performed using the Poor Man's PCR protocol (Schuelke 2000) as previously described (Spigleret al. 2008). The 15-μl reactions included 0.4 units HotStarTaq Polymerase (Qiagen) with template DNA at a concentration of 0.002 ng/μl, and 1.5 mm MgCl supplied by 10× PCR buffer (Qiagen), 0.67 mm dNTPs (Applied Biosystems), 120 nM sequence-specific forward primer with M13(-21) 5′ universal sequence tag, and 500 nM each of sequence-specific reverse primer and fluorescence-labeled M13(-21) universal primer (Applied Biosystems). An initial 15-min incubation period at 94° was followed by 30 cycles of melting at 94°, annealing at 59°, and elongation at 72° with 45 sec at each step. A second round of 15 cycles utilizing a lower annealing temperature of 53° allowed annealing of the fluorescence-tagged M13(-21) universal primer to the M13(-21) sequence tags on the PCR products produced in the previous 30 cycles, generating fluorescence-labeled PCR products for detection in capillary electrophoresis (Schuelke 2000). Prior to capillary electrophoresis on an ABI 3730 DNA Analyzer (Applied Biosystems), 1 μl of each reaction was mixed with 0.5 μl LIZ500 standard and 8.5 μl Hi-Di formamide (Applied Biosystems). Electrophoretic data were visualized and products assessed using GeneMapper (Applied Biosystems).
Map construction:
Because F. chiloensis is an octoploid, primers may amplify products on more than one homeologous chromosome. Thus, we treated all PCR products (“markers”) and the two phenotypic sex traits as single-dose markers (see Wuet al. 1992) and evaluated their fit according to expected Mendelian segregation ratios of either 1:1, if present in only one parent, or 3:1, if present in both parents. Because of the number of markers evaluated, we retained only markers that fitted expected ratios at P ≥ 0.0001. Most (84%) markers retained for mapping fitted at P ≥ 0.05. We recorded markers that deviated significantly from expected segregation ratios at P ≤ 0.05. To identify which products of a given primer pair represented alleles at a single locus, we first mapped products from a given primer pair in JoinMap 4.0 (VanOoijen 2006). We then retained one member of each allelic pair for creating the full map, consistent with the single-dose marker approach of Wuet al. (1992) (also see Spigleret al. 2010).
To create separate maps of HG VI for the male and female parents in F. chiloensis, we used a pseudobackcross strategy (Grattapaglia and Sederoff 1994). Both maps were created in JoinMap 4.0 as a “cross pollinators” (CP) family type for an outbred full-sib family cross. We initially excluded markers from the analysis that were missing data for >25% of individuals in the mapping population, and four individuals were excluded because they were missing >50% of marker data. Initial linkage groups were inferred at LOD 6, and we subsequently added ungrouped and previously excluded markers to the existing groups at LOD > 4 using the strongest cross link (SCL) values. For pairs of markers from a given primer pair found to be allelic in the step before mapping, we retained only one marker for mapping once the markers were assigned groups. We determined marker order and map distance using the Kosambi mapping function and the default mapping parameters in JoinMap (minimum LOD threshold 1.0, recombination threshold of 0.40, and jump threshold of 5.0). Graphical maps were generated in MapChart 2.1 (Voorrips 2002). Once linkage groups in HG VI in F. chiloensis were assembled, we named them on the basis of LG-specific markers shared with the F. virginiana map in Spigleret al. (2010). Thus, LG designations presented here for F. chiloensis represent hypotheses about homeology with F. virginiana.
Comparative analysis of macrosynteny:
To characterize the sex chromosome in F. chiloensis, we compared the LG carrying the region(s) controlling male and female function in F. chiloensis to (1) the proto-sex chromosome identified in subdioecious F. virginiana (Spigleret al. 2010) and to (2) the homeolog from a map based on a cross between two hermaphroditic diploid congeners [F. vesca and F. nubicola, hereafter “FV×FN” (Sargentet al. 2008)]. For this comparison, we included only those DNA markers in the FV×FN LG 6 map (data kindly provided by D. Sargent) that were shared with both the F. chiloensis map in this study and the F. virginiana HG VI map from Spigleret al. (2010). More stringent mapping parameters were used in both Sargentet al. (2006) and Spigleret al. (2010) to determine marker ordering. We therefore re-evaluated marker order of the F. chiloensis linkage group housing male and female function genes using JoinMap with similarly stringent parameters (a recombination threshold of 0.35, LOD threshold of 3.0, and jump threshold of 3.0). Marker order did not change using these parameters, and we present the map derived from our original analysis (see above).
RESULTS
Sex expression:
Of the 84 progeny that flowered in our mapping population, 45 (54%) were male sterile and 39 (46%) were male fertile. Seventy-six percent of the offspring scored for fruit set were female fertile and 24% were female sterile. Thus, when scored qualitatively, male function segregated in a 1:1 ratio (χ2 = 0.76; P = 0.38), and female function segregated in a 3:1 ratio (χ2 = 0.02; P = 0.90). Overall, 21% were both male fertile and female fertile (“hermaphrodite”), 55% were male sterile and female fertile (“female”), and 24% were male fertile and female sterile (“male”). These results confirm the dominance of male sterility over fertility and the recessivity of female sterility vs. fertility. The segregation ratios are compatible with a one-locus (Fm and hm) or a two-locus (AG|ag and aG|ag) interpretation with the maternal parent having AG and ag in coupling and the paternal parent having aG and ag in coupling (see Table 1). We cannot distinguish between these two models since no neuters definitively indicating recombination were observed; note that that some other recombinant types would be phenotypically indistinguishable from nonrecombinants (Table 1). A test to distinguish between allelism at a single locus vs. two linked loci with suppressed recombination will probably require a mapping population many-fold larger than the current one (Liuet al. 2004; Telgmann-Rauberet al. 2007).
Hypothesized genotypes at sex loci of F. chiloensis map cross parents and the genotypes and phenotypes of their offspring
Parental genotype . | Gametes (nonrecombinant)a . | Offspring fertility . | Progeny phenotype . | Gametes (recombinant)a . | Offspring fertility . | Progeny phenotype . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Maternal . | Paternal . | Maternal . | Paternal . | Female . | Male . | Maternal . | Paternal . | Female . | Male . | ||
| AG|ag | aG|ag | AG | aG | + | − | Female | Ag | aG | + | − | Female |
| ag | ag | − | + | Male | aG | ag | + | + | Hermaphrodite | ||
| AG | ag | + | − | Female | Ag | ag | − | − | Neutera | ||
| ag | aG | + | + | Hermaphrodite | aG | aG | + | + | Hermaphrodite | ||
Parental genotype . | Gametes (nonrecombinant)a . | Offspring fertility . | Progeny phenotype . | Gametes (recombinant)a . | Offspring fertility . | Progeny phenotype . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Maternal . | Paternal . | Maternal . | Paternal . | Female . | Male . | Maternal . | Paternal . | Female . | Male . | ||
| AG|ag | aG|ag | AG | aG | + | − | Female | Ag | aG | + | − | Female |
| ag | ag | − | + | Male | aG | ag | + | + | Hermaphrodite | ||
| AG | ag | + | − | Female | Ag | ag | − | − | Neutera | ||
| ag | aG | + | + | Hermaphrodite | aG | aG | + | + | Hermaphrodite | ||
Gametes are represented with and without recombination between the sex loci. Recombination in the maternal parent results in gametes with allelic combinations for the sex loci that differ from the parental genotype, whereas recombination in the paternal parent does not.
Only neuter offspring are phenotypically distinguishable as recombinants.
Hypothesized genotypes at sex loci of F. chiloensis map cross parents and the genotypes and phenotypes of their offspring
Parental genotype . | Gametes (nonrecombinant)a . | Offspring fertility . | Progeny phenotype . | Gametes (recombinant)a . | Offspring fertility . | Progeny phenotype . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Maternal . | Paternal . | Maternal . | Paternal . | Female . | Male . | Maternal . | Paternal . | Female . | Male . | ||
| AG|ag | aG|ag | AG | aG | + | − | Female | Ag | aG | + | − | Female |
| ag | ag | − | + | Male | aG | ag | + | + | Hermaphrodite | ||
| AG | ag | + | − | Female | Ag | ag | − | − | Neutera | ||
| ag | aG | + | + | Hermaphrodite | aG | aG | + | + | Hermaphrodite | ||
Parental genotype . | Gametes (nonrecombinant)a . | Offspring fertility . | Progeny phenotype . | Gametes (recombinant)a . | Offspring fertility . | Progeny phenotype . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Maternal . | Paternal . | Maternal . | Paternal . | Female . | Male . | Maternal . | Paternal . | Female . | Male . | ||
| AG|ag | aG|ag | AG | aG | + | − | Female | Ag | aG | + | − | Female |
| ag | ag | − | + | Male | aG | ag | + | + | Hermaphrodite | ||
| AG | ag | + | − | Female | Ag | ag | − | − | Neutera | ||
| ag | aG | + | + | Hermaphrodite | aG | aG | + | + | Hermaphrodite | ||
Gametes are represented with and without recombination between the sex loci. Recombination in the maternal parent results in gametes with allelic combinations for the sex loci that differ from the parental genotype, whereas recombination in the paternal parent does not.
Only neuter offspring are phenotypically distinguishable as recombinants.
Another result was that, among the male-sterile progeny, 46% produced exclusively white vestigial anthers, while 54% consistently or occasionally produced small yellow anthers with a small amount of immature pollen in their indehiscent anther sacs (χ2 = 0.56; P = 0.33).
The map of F. chiloensis HG VI:
Ninety-one percent of the primer pairs amplified product(s) in the F. chiloensis mapping population. Of the 300 products from these 51 primer pairs, 189 met our criteria for consideration for map construction (112 for the maternal map and 107 for the paternal map).
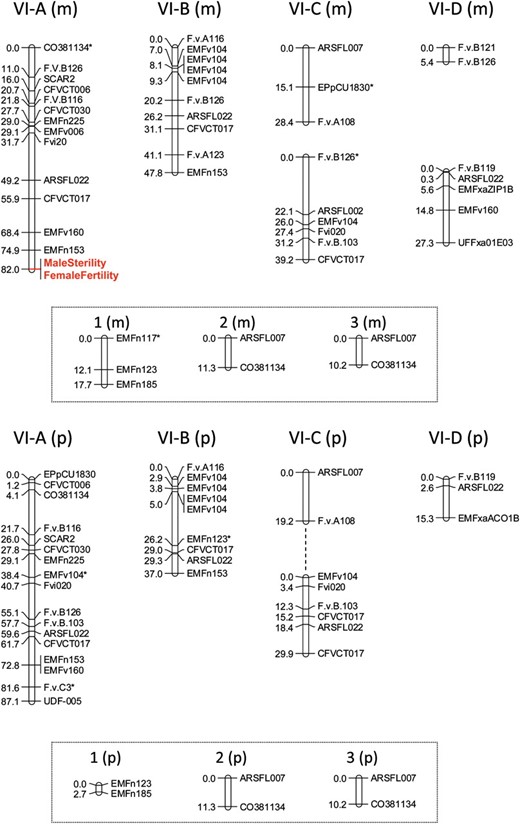
The maternal map of HG VI (Figure 1, top) included 46 PCR-based markers (from 73 PCR amplification products of which 54 were identified as members of allelic pairs), plus the phenotypic sex-function markers, and formed nine LGs. We were able to assemble six LGs to represent the four homeologous chromosomes of HG VI (the homeologous group to which the sex-determining chromosomes in F. virginiana also belong; see Spigleret al. 2010) on the basis of the presence of certain markers. For example, four main independent LGs were identified on the basis of possessing a copy of the F.v.B126 locus. These four LGs ranged in size from ∼5 to 82 cM, with between 2 and 14 markers per group and an average marker spacing of 7.09 cM (±5.13 SD). Importantly, the male sterility and female fertility factors mapped to one of these four LGs with no detectable recombinants between them; we thus label the sex-function genes as a single marker in the map (Figure 1, top).
Parental linkage maps of linkage groups belonging to homeologous group VI in F. chiloensis. Linkage groups identified as distinct from one another (see results) are given names according to the homeologous group (VI) and a letter (A–D). Additional linkage groups beyond those four major groups are named using Arabic numbers. Maternal linkage groups (top) are denoted by “m”; paternal linkage groups (bottom) are denoted by “p.” Linkage groups with similar names between the parental maps are putative homeologs. Associations between pairs of linkage groups within the parental maps were identified either through SCL values between markers in the two groups at LOD > 4 (indicated by dashed line) or through comparison to the putative homeolog in the other parent map. Phenotypic trait markers representing the putative sex-determining loci are indicated in enlarged (red) font. Marker names denoted by an asterisk had segregation ratios that deviated significantly from the expected (P < 0.05).
The paternal map of HG VI (Figure 1, bottom) has 43 distinct markers (64 products, including 42 members of allelic pairs) across eight linkage groups. Using the approach described above for the maternal map, we were able to assemble five LGs to represent the four HG VI homeologous chromosomes. Specifically, ARSFL22 was found in each of four main LGs. LGs in the paternal map ranged in size from ∼3 to 87 cM. LGs had between 2 and 17 markers, with an average marker spacing of 6.08 cM (±5.79 SD).
Most markers in each parental map were parent-specific, but five were shared between the parent maps. Using this information and comparisons of macrosynteny between LGs in the two maps, we were able to identify homeologous chromosomes in the two parental maps. We could similarly indentify putatively LG-specific markers in the F. virginiana map (see Spigleret al. 2010), allowing us to identify chromosomes in the HG with LGs in the F. chiloensis map (Figure 1). For example, because F.v.A108 is LG-specific in both the F. virginiana and F. chiloensis mapping populations, we can identify LG VI-C in F. chiloensis. Similarly, SCAR2 mapped to LG VI-A and F.v.B119 to LG VI-D in F. virginiana, and we therefore named the F. chiloensis LGs containing these markers VI-A and VI-D, respectively. The remaining major linkage group could thus be deduced to be VI-B. These data confirm that the F. chiloensis sex chromosome is in HG VI, but suggest that it is a different homeolog (VI-A) from that in F. virginiana (VI-C).
Comparative analysis of macrosynteny:
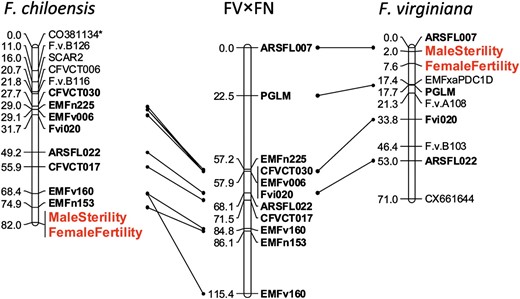
Comparison of the map of the sex chromosome in F. chiloensis (VI-A) with that of the proto-sex chromosome of F. virginiana (VI-C) and the homeologous autosome in a hermaphroditic diploid cross (LG 6) reveals substantial macrosynteny across these three LGs (Figure 2). Specifically, the maternal map of the F. chiloensis sex-determining chromosome (VI-A) shares seven markers with LG 6 in the diploid FV×FN cross and two with the maternal map of VI-C, the proto-sex chromosome of F. virginiana, and in both cases all of these markers are in the same order. A further indication that the orientation of the F. chiloensis sex chromosome is correct is our finding of the SCAR2 marker at the top of VI-A; this marker has been shown to cosegregate with the seasonal flowering locus (Albaniet al. 2004), which has been mapped to the top of LG 6 in the diploid FV×FN map (Sargentet al. 2004).
Comparison of the F. chiloensis putative sex chromosome (VI-A) to previously published maps of a diploid Fragaria (FV×FN) autosomal homeolog (LG 6) and the proto-sex chromosome identified in F. virginiana (VI-C). Lines connect primer pairs shared between the respective octoploid sex chromosomes and the diploid reference homeolog linkage group. Phenotypic trait markers representing the putative determining sex loci are indicated in enlarged (red) font. Marker names denoted by an asterisk had segregation ratios that deviated significantly from the expected (P < 0.05).
Despite the observed macrosynteny, there is also clear evidence that the location of the sex-determining region differs between F. chiloensis and F. virginiana. In F. chiloensis, male sterility is linked to the markers EMFn153 (at a LOD of 15.01) and EMFv160 (at a LOD of 9.73), both of which clearly map to the bottom of the homeologous LG 6 in the diploid (Sargentet al. 2004). Furthermore, SCL values found for male sterility to a marker outside of the group are very weak (LOD of 1.9). This contrasts with the location of the sex-determining genes in F. virginiana among markers at the top of LG VI-C. In the F. virginiana mapping population, male sterility was linked to markers ARSFL007 and F.v.A108 at LOD scores of 44.62 and 18.42, respectively, and ARSFL007 clearly maps to the top of the diploid LG 6 (Spigleret al. 2010). ARSFL007 and F.v.A108 also are linked in F. chiloensis at a similar distance as in F. virginiana, but male sterility clearly does not map between them. Since differences in population size can affect mapping results, we randomly reduced the original F. virginiana mapping population to a size equivalent to that used in our study of F. chiloensis. We found that the LOD scores for associations between ARSFL007, F.v.A108, and male sterility remain high (>10) and are similar to the LOD scores found here for F. chiloensis, confirming that the difference in position of male sterility in the two species is not a function of mapping population size but reflects a true difference.
DISCUSSION
The main interesting result of our new genetic linkage study is that, although the dominance relations of the loci affecting male and female function in F. chiloensis are similar to those in F. virginiana and the chromosome carrying the sex-determining genes in both species belongs to the same homeologous group (HG VI), the locations of the sex-determining region on the chromosomes differ. Thus, the two species may not share the same sex-determining region as has previously been assumed (Ahmadi and Bringhurst 1991).
The segregation of two phenotypic subclasses (vestigial vs. yellow anthers) of male-sterile progeny in this cross was not observed in F. virginiana (T.-L. Ashman, personal observation) and raises the possibility that additional genetic factors influence sexual expression in F. chiloensis. For example, ag and aG chromosomal regions from the paternal parent that segregate 1:1 in the female progeny may have different phenotypic effects not only in F. chiloensis male-fertile individuals by conferring complete (e.g., aagg; males) or partial female sterility (e.g., aaGg; hermaphrodites), but also in F. chiloensis male-sterile individuals with respect to anther development (e.g., AaGG vs. AaGg females) (Table 1). This is interesting because evolutionary theory predicts that alleles that promote male function will become linked to the region controlling female sterility (“g” in the notation used here) and that in the early evolution of sex chromosomes such alleles may be present (and expressed) in both sexes (reviewed in Charlesworth 2008). An alternative is that there is developmental leakiness of male sterility (A allele) in F. chiloensis. Developmental leakiness, or plasticity, in sexual expression could be due to variable methylation of sex-determining genes (Gorelick 2003) or transposons that can accumulate in nonrecombining regions of sex chromososmes (Zhanget al. 2008). Finally, it is also possible that the anther phenotypes of females are the result of alleles segregating at loci in other regions of the genome that influence the anther developmental pathway. Additional studies will be required to differentiate between these possibilities.
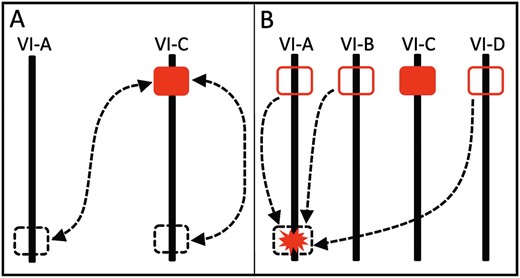
Do the two octoploid species share the same ancestral sex-determining region? Our identification of the most tightly linked SSR markers and inferences that their chromosomal positions differ might suggest that they do not. However, a single origin for the sex-determining region in F. chiloensis and F. virginiana (or at least for an initial dominant male sterility mutation) in a common ancestor of both species is not excluded because a chromosomal translocation might have moved the sex-determining region from the top of one HG VI chromosome to the bottom of another (Figure 3A). Indeed, an SSR marker close to sex in F. virginiana has been involved in a translocation to another chromosome (Spigleret al. 2010). Translocations of existing sex-determining regions to form neo-sex chromosomes occur in systems with both young and old sex chromosomes (see Introduction). Furthermore, a translocation could account for the apparent colocalization of loci for male and female function in F. chiloensis (i.e., absence of recombination between them), as translocations and inversions in sex regions are a common mechanism of recombination suppression (reviewed in Bergero and Charlesworth 2009) and sex chromosome formation (reviewed in Charlesworth and Charlesworth 1978).
Scenarios by which the F. chiloensis sex chromosome may have evolved. Solid red rectangles represent a sex-determining region homeologous to that in F. virginiana, while outlined red rectangles represent the gene region homeologous to this sex-determining region without sterility mutations. Dashed lines show translocation events while dashed rectangles represent the location of sex-determining genes either pre- or post-translocation, depending on the direction of arrows. The red starburst represents a gene region on LG VI-A with a novel sterility mutation. (A) Intra- and interchromosomal translocation events of an ancestral sex-determining region to or from the top of LG VI-C. (B) Possible interchromosomal translocations of F. virginiana sex-determining region homeologs to the bottom of LG VI-A. Alternatively, novel sterility mutations in a gene region located at the bottom of LG VI-A could have led to the independent formation of a sex-determining region.
An alternative is that sex-determining regions evolved independently in the two species. Independent dominant mutations must have caused male sterility in F. virginiana and F. chiloensis, either in different genes or in the duplicate copies of the same gene, but in either case on initially autosomal members of HG VI (Figure 3B). A translocation would be involved only if the mutations occurred in homeologus gene copies. Linkage group 6 in diploid Fragaria carries many genes involved in reproductive function, including self-incompatibility (Boskovicet al. 2010), flower size (Sargent 2005), and pollen development (T.-L. Ashman, P. Jaisiwal, A. Liston, M. Hanumappa and J. Elser, unpublished data). The pollen development pathway, in particular, is complex (reviewed in Borget al. 2009 and Borg and Twell 2010), and thus mutations in different genes could potentially cause dominant loss of function (e.g., ASHR3; Thorstensenet al. 2008). The four copies of chromosome 6 in octoploid Fragaria increase the opportunities for mutations in reproductive function genes. Gene duplication may also facilitate shifts in the master regulator of a sex-determining developmental pathway (see figure 1 in Schartl 2004). Additional work in these and other closely related sexually polymorphic species is underway to fully to reveal the evolutionary history of sex chromosomes in Fragaria.
Footnotes
Communicating editor: D. Charlesworth
Acknowledgements
We thank C. E. Ashman, A. Funk, K. Lewers, B. McTeague, and E. York for assistance; D. Sargent for files of the diploid map; the members of the Ashman lab and the University of Pittsburgh's Howard Hughes Medical Institute's Undergraduate Science Education Program for discussion; and K. Lewers, D. Charlesworth, and two anonymous reviewers for helpful comments on a previous version of the manuscript. This work was supported in part by the National Science Foundation (DEB 0449488 and 1020523) and the Howard Hughes Medical Institute (grant 52005906).
References
Ahmadi, H., and R. S. Bringhurst,
Albani, M. C., N. H. Battey and M. J. Wilkinson,
Ashley, M. V., J. A. Wilk, S. M. N. Styan, K. J. Craft, K. L. Jones et al.,
Ashman, T.,
Bachtrog, D.,
Bachtrog, D., and B. Charlesworth,
Bergero, R., and D. Charlesworth,
Bergero, R., A. Forrest, E. Kamau and D. Charlesworth,
Bishop, E. J., R. B. Spigler and T. L. Ashman,
Borg, M., and D. Twell,
Borg, M., L. Brownfield and D. Twell,
Boskovic, R. I., D. J. Sargent and K. R. Tobutt,
Bringhurst, R. S.,
Bull, J. J.,
Catling, P. M., and S. Porebski,
Charlesworth, B., and D. Charlesworth,
Charlesworth, D.,
Charlesworth, D.,
Charlesworth, D.,
Chen, C. X., Q. Y. Yu, S. B. Hou, Y. J. Li, M. Eustice et al.,
Cnaani, A., B. Y. Lee, N. Zilberman, C. Ozouf-Costaz, G. Hulata et al.,
Cunado, N., R. Navajas-Perez, R. de la Herran, C. R. Rejon, M. R. Rejon et al.,
Desfeux, C., S. Maurice, J.-P. Henry, B. Lejeune and P.-H. Gouyon,
Filatov, D. A.,
Fraser, J. A., and J. Heitman,
Gorelick, R.,
Grattapaglia, D., and R. Sederoff,
Hancock, J. F. J., Jr., and R. S. Bringhurst,
Hancock, J. F., S. Serce, C. M. Portman, P. W. Callow and J. J. Luby,
Hokanson, K. E., M. J. Smith, A. M. Connor, J. J. Luby and J. F. Hancock,
Howell, E. C., S. J. Armstrong and D. A. Filatov,
Lerceteau-Köhler, E., G. Guérin, F. Laigret and B. Denoyes-Rothan,
Liu, Z., P. H. Moore, C. M. Ackerman, M. Ragiba Q. Yu, et. al.,
Ma, H., P. H. Moore, Z. Y. Liu, M. S. Kim, Q. Y. Yu et al.,
Marguerit, E., C. Boury, A. Minicki, M. Donnart, G. Butterlin et al.,
Mrackova, M., M. Nicolas, R. Hobza, I. Negrutiu, F. Moneger et al.,
Navajas-Perez, R., R. de la Herran, G. L. Gonzalez, M. Jamilena, R. Lozano et al.,
Njuguna, W.,
Ogata, M., Y. Hasegawa, H. Ohtani, M. Mineyama and I. Miura,
Reamon-Buettner, S. M., J. Schondelmaier and C. Jung,
Renner, S. S., and R. E. Ricklefs,
Ross, J. A., J. R. Urton, J. Boland, M. D. Shapiro and C. L. Peichel,
Rousseau-Gueutin, M., E. Lerceteau-Köhler, L. Barrot, D. J. Sargent, A. Monfort et al.,
Rousseau-Gueutin, M., A. Gaston, A. Aïnouche, M. L. Aïnouche, K. Olbricht et al.,
Sargent, D. J.,
Sargent, D. J., T. M. Davis, K. R. Tobutt, M. J. Wilkinson, N. H. Battey et al.,
Sargent, D. J., J. Clarke, D. W. Simpson, K. R. Tobutt, P. Arús et al.,
Sargent, D., G. Cipriani, S. Vilanova, D. Gil-Ariza, P. Arús et al.,
Sargent, D. J., F. Fernandéz-Fernandéz, J. J. Ruiz-Rojas, B. G. Sutherland, A. Passey et al.,
Schartl, M.,
Schuelke, M.,
Sondur, S. N., R. M. Manshardt and J. I. Stiles,
Spigler, R. B., K. S. Lewers, D. S. Main and T.-L. Ashman,
Spigler, R., K. Lewers, A. Johnson and T. Ashman,
Staudt, G.,
Staudt, G.,
Steinemann, M., and S. Steinemann,
Telgmann-Rauber, A., A. Jamsari, M. S. Kinney, J. C. Pires and C. Jung,
Testolin, R., W.G. Huang, O. Lain, R. Messina, A. Vecchione et al.,
Thorstensen, T., P. E. Grini, I. S. Mercy, V. Alm, S. Erdal et al.,
Uno, Y., C. Nishida, Y. Oshima, S. Yokoyama, I. Miura et al.,
Uno, Y., C. Nishida, S. Yoshimoto, M. Ito, Y. Oshima et al.,
Van Ooijen, J. W.,
Voorrips, R. E.,
Westergaard, M.,
Wu, K. K., W. Burnquist, M. E. Sorrells, T. L. Tew, P. H. Moore et al.,
Wu, X., J. Wang, J.-K. Na, Q. Yu, R. C. Moore et al.,
Yu, Q., S. Hou, F. A. Feltus, M. R. Jones, J. E. Murray et al.,
Yu, Q., R. Navajas-Pérez, E. Tong, J. Robertson, P. H. Moore et al.,
Zhang, W., X. Wang, Q. Yu, R. Ming and J. Yang,