-
PDF
- Split View
-
Views
-
Cite
Cite
William S Talbot, Elizabeth S Egan, Michael A Gates, Charline Walker, Bonnie Ullmann, Stephan C F Neuhauss, Charles B Kimmel, John H Postlethwait, Genetic Analysis of Chromosomal Rearrangements in the cyclops Region of the Zebrafish Genome, Genetics, Volume 148, Issue 1, 1 January 1998, Pages 373–380, https://doi.org/10.1093/genetics/148.1.373
Close - Share Icon Share
Abstract
Genetic screens in zebrafish have provided mutations in hundreds of genes with essential functions in the developing embryo. To investigate the possible uses of chromosomal rearrangements in the analysis of these mutations, we genetically characterized three gamma-ray induced alleles of cyclops (cyc), a gene required for development of midline structures. We show that cyc maps near one end of Linkage Group 12 (LG 12) and that this region is involved in a reciprocal translocation with LG 2 in one gamma-ray induced mutation, cyc b213. The translocated segments together cover approximately 5% of the genetic map, and we show that this rearrangement is useful for mapping cloned genes that reside in the affected chromosomal regions. The other two alleles, cycb16 and cycb229, have deletions in the distal region of LG 12. Interestingly, both of these mutations suppress recombination between genetic markers in LG 12, including markers at a distance from the deletion. This observation raises the possibility that these deletions affect a site required for meiotic recombination on the LG 12 chromosome. The cycb16 and cycb229 mutations may be useful for balancing other lethal mutations located in the distal region of LG 12. These results show that chromosomal rearrangements can provide useful resources for mapping and genetic analyses in zebrafish.
THE zebrafish (Danio rerio) has become an important model organism for the study of vertebrate biology (Kimmel 1989; Driever et al. 1994; Eisen 1996; Felsenfeld 1996; Grunwald 1996; Holder and McMahon 1996). Large-scale mutagenesis screens have provided mutations in hundreds of genes essential for normal development, physiology, and behavior (Riley and Grunwald 1995; Driever et al. 1996; Gaiano et al. 1996; Haffter et al. 1996; Henion et al. 1996). Moreover, the optical clarity and accessibility of the embryo allow detailed phenotypic characterization with techniques such as cellular transplantation and lineage tracing (Ho and Kane 1990; Hatta et al. 1991; Melby et al. 1996). Thus, the cellular functions of genes defined by mutations in zebrafish can be studied in great detail. Molecular genetic analysis of these mutant loci is now an important goal, and development of genetic tools and genomic resources that facilitate the cloning of these genes is therefore a high priority (Postlethwait et al. 1994; Johnson et al. 1996; Knapik et al. 1996; Postlethwait and Talbot 1997).
Chromosomal rearrangements induced by gammarays and X-rays, including deletions, duplications, translocations, and inversions are important tools in the genetic analysis of a number of model organisms. In Drosophila, for example, chromosomal rearrangements are used to distinguish amorphic from hypomorphic alleles, to balance lethal and sterile mutations, to map mutations, to investigate gene dosage effects, and to identify mutated genes through breakpoint mapping (Ashburner 1989). By comparison, little has been done to characterize gamma-ray induced mutations in zebrafish, although gamma-ray induced deficiencies have been used in zebrafish to establish null phenotype (Talbot et al. 1995; Schier et al. 1997; Halpern et al. 1997) and to remove chromosomal segments containing specific cloned genes (Fritz et al. 1996).
We set out to determine the genetic nature of three gamma-ray induced mutations involving cyclops (cyc), a gene essential for development of the embryonic midline (Hatta et al. 1991). We found that two of these alleles, cycb16 and cycb229, have deletions in the distal region of Linkage Group 12 (LG 12), and that both suppress recombination between markers in a segment of LG 12, thereby functioning as balancer chromosomes for this chromosomal region. The third allele, cycb213, is a reciprocal translocation between LG 12 and LG 2. These results suggest that the systematic collection and analysis of gamma-ray induced mutations in other parts of the genome will provide important tools for genetic analysis in zebrafish.
MATERIALS AND METHODS
Fish strains: The AB strain (Chakrabarti et al. 1983) was used in the screens that identified cycb213 and cycb229. Other strains used to produce hybrids for mapping include DAR, Tü, and TL (Postlethwait et al. 1994; Haffter et al. 1996).
Nomenclature: We followed previous linkage group designations (Postlethwait et al. 1994; Johnson et al. 1996). Each linkage group corresponds to a different chromosome because each has been assigned a centromere (Johnson et al. 1996).
Following guidelines for Drosophila rearrangements, the b213 reciprocal translocation is described as T(LG2;LG12)b213, and the two elements of the translocation are termed T(LG2; LG12)b213, 2P12D (for the rearranged chromosome with the centromere-proximal segment of LG 2 and distal segment of LG 12) and T(LG2;LG12)b213, 12P2D. Segregation of these rearranged chromosomes and their normal order counterparts results in euploid and aneuploid meiotic products (see Figure 7). For convenience of discussion, we refer to the haploid genotype with a normal order LG 2 and a T(LG2;LG12)b213, 12P2D chromosome as cycb213 and the genotype with a normal order LG 12 and a T(LG2;LG12)b213, 2P12D chromosome as necb213, referring to the cyclopic and necrotic phenotypes characteristic of aneuploid haploid embryos with these genotypes. We emphasize that the cycb213 and necb213 phenotypes result from the loss and duplication of chromosomal segments, not individual genes.
Isolation of new cyclops alleles: The identification of cycb16 and cycm294 has been described (Hatta et al. 1991; Schier et al. 1996). cycb213 was identified in a screen for gamma-ray induced mutations causing morphological defects in the embryo (Kimmel 1989). The cycb213 mutation fails to complement cycb16. The cycb229 mutation was identified in a screen for gamma-ray induced mutations that fail to complement cycb16. To examine the possibility that cycb229 was an inadvertent reisolate of cycb16, we compared alleles of PCR-based genetic markers present on the two mutant chromosomes. The cycb16 and cycb229 chromosomes have different alleles of a simple sequence length polymorphism (SSLP) marker (z1400) in the region where recombination is suppressed by both mutations (see Figure 2), suggesting that cycb229 is a newly generated cyc allele. The observation that cycb229 recombines more frequently with proximal LG 12 markers, e.g., z3801, (see Table 1) than cycb16 provides further evidence for the independent origin of the two alleles.
Genetic mapping and markers: Generation of haploid embryos, genomic DNA preparation, and PCR conditions have been described (Postlethwait et al. 1994; Johnson et al. 1996). Polymorphisms were detected either by agarose gel electrophoresis and ethidium bromide staining or by acrylamide gel electrophoresis of 32P-labelled PCR products.
The LG 12 markers shown in Figure 1C were mapped in at least one of two haploid mapping panels. One panel was produced from 48 haploid progeny of a TL/Tü female and the other was produced from 48 haploid progeny of an AB/Tü female. Maps were compiled with Map Manager (K. Manley and R. Cudmore, http://mcbio.med.buffalo.edu/mapmgr.html). Maps produced from these two panels were integrated with each other by reference to markers scored in both panels.
Primers for SSLP markers (Knapik et al. 1996) were obtained commercially (Research Genetics, Birmingham, AL). The primers used to amplify ptc1 (Concordet et al. 1996) were PTCF1 (5′-AGCAAGGAGCTACGCTACAC-3′) and PTCR1 (5′-GCAGGGGAAAAGCTTATCAA-3′). The primers for other genes shown in Figure 7 will be described elsewhere (J.H.P. et al., in preparation). The randomly amplified polymorphic DNA (RAPD) markers 18AF.1190, 15T.750, and 12R.360 (Postlethwait et al. 1994; Johnson et al. 1996; Halpern et al. 1997) were converted to sequence-tagged site (STS) markers by deriving locus-specific primers from the sequences of the cloned RAPD fragments. The primers for the 18AF.1190 STS were 18AF.1190F (5′-CCCTCTGCACAGAACTGAAACCTC-3′) and 18AF.1190R (5′-CCGTTTCCTGTGAAGACAGGAAG-3′). The primers for the 15T.750 STS were 15T.750F (5′-CTGTCT GGAGAAAGTCTTATTTG-3′) and 15T.750R (5′-GGATGC CACTGGTACTAATTGATA-3′). The primers for the 12R.360 STS were 12R.360F (5′-ACAGGTGCGTCCAATAGCTCC-3′) and 12R.360R (5′-ACAGGTGCGTGATCAAATGTT-3′). These loci were scored as STS markers in some experiments (such as those shown in Figures 1, 3, 5, and 6) and as RAPDs in others (including some experiments shown in Table 1).
RESULTS
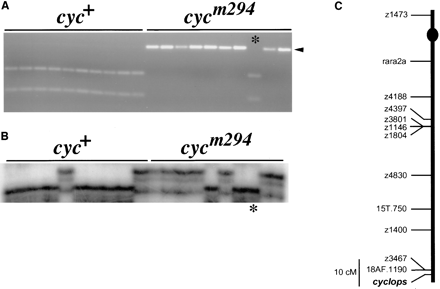
Genetic mapping of cyclops: Previous work demonstrated that cyclops is linked to randomly amplified polymorphic DNA (RAPD) markers in the distal region of Linkage Group 12 (LG 12; Postlethwait et al. 1994). This assignment employed cycb16, which we show below to be a rearrangement that suppresses recombination over part of LG 12 and therefore obscures the exact position of cyc. To avoid this difficulty, we analyzed LG 12 markers in mapping crosses constructed with cycm294, which was induced with N-ethyl-N-nitrosourea (ENU) and thus is likely to be a point mutation (Schier et al. 1996). Scoring LG 12 markers, including RAPDs, simple sequence length polymorphisms (SSLPs), and sequence-tagged sites (STSs) derived from cloned RAPDs, in individual haploid progeny of DAR x cycm294 F1 females confirmed the assignment of cyc to LG 12 (Figure 1). The cycm294 mutation is linked to 18AF.1190 (1 recombinant among 44 haploid individuals), a RAPD that has been cloned and converted into an STS (Figure 1A). Analysis of markers proximal to 18AF.1190, such as SSLP z1400, in the same cycm294 mapping crosses indicated that cyc is distal to 18AF.1190 (Figure 1, B and C). LG 12 markers were also scored in haploid embryos from wild-type, i.e., cyc+, reference mapping panels (Postlethwait et al. 1994; Johnson et al. 1996; this work, see materials and methods). Distances between LG 12 markers do not differ significantly in cyc+ and cycm294 mapping panels, indicating that cycm294 maps as a point mutation or other small lesion, as would be expected for an ENU-induced mutation. For example, there were 6 recombinants between markers 18AF.1190 and z1400 among 44 individuals in a wild-type mapping panel and 4 recombinants among 44 individuals in a cycm294 mapping panel (Figure 1 and Table 1).
cycb16 and cycb229 delete markers near cyc: The cycb16 and cycb229 mutations are gamma-ray induced cyc alleles that segregate as Mendelian recessives (Hatta et al. 1991; Halpern et al. 1997; Table 2, this work). To investigate the possibility that these mutations involve deletions in
Genetic mapping of cyclops. (A) The marker 18AF.1190 is linked to cyc. Ten wild-type and 10 cycm294 haploid siblings were analyzed with an STS marker derived from the 18AF.1190 RAPD. An Mnl I site polymorphism reveals that the 18AF.1190 locus is linked to cyc, and one recombinant (*) is shown. This was the only recombinant identified among 44 haploid progeny of a DAR x cycm294 female. The uncleaved 18AF.1190 fragment is 360 bp. The arrowhead marks the 300-bp fragment that is linked in coupling to the mutant chromosome; the other allele is marked by 190-bp and 110-bp Mnl I fragments. (B) The same 20 embryos were analyzed with the SSLP marker z1400. Five recombinants are shown, including the recombinant identified with 18AF.1190 (*) in A. This places z1400 and 18AF.1190 on the same side of cyc. Five recombinants for z1400 and cyc were identified among 44 haploid progeny of the DAR x cycm294 female. (C) Genetic map of Linkage Group 12, showing 18AF.1190, z1400, and cyc. The markers shown were scored in a wild-type, i.e., cyc+, haploids, and a map was generated (see materials and methods). The position of cyc was inferred from the analysis of z1400 and 18AF.1190 in the cycm294 mapping cross. The centromere location (dot) has been reported previously (Johnson et al. 1996). The map order shown is consistent with previous maps (Postlethwait et al. 1994; Johnson et al. 1996; Knapik et al. 1996).
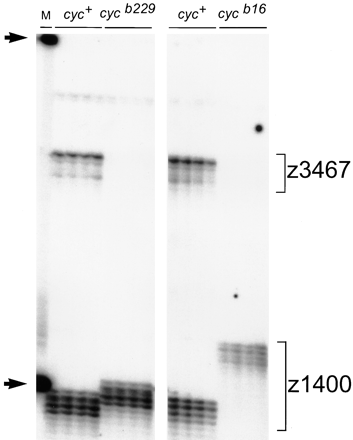
the vicinity of cyc, we assayed LG 12 markers on genomic DNA isolated from cycb16 and cycb229 haploid embryos and their wild-type siblings. Figure 2 shows that one of the markers nearest to cyc, the SSLP z3467, amplifies from the wild-type samples, but not from cycb16 or cycb229 DNA. This result, together with our observations that 18AF.1190 and other nearby markers give similar results (data not shown), indicates that both cycb16 and cycb229 contain deletions in the vicinity of cyc. The SSLP z1400, which lies about 10 cM from z3467 and 18F.1190 (Figure 1C), amplifies from both wild-type and mutant samples (Figure 2), indicating that neither cycb16 nor cycb229 involves a deletion extending beyond this marker.
cyc b16 and cyc b229 suppress recombination on LG 12: During the analysis of cycb16 and cycb229, we noted
Genetic distance between cyc and LG 12 markers in cyc mutant mapping crosses
| . | Percent recombination with cyc (No. rec./total) . | |||
|---|---|---|---|---|
| cyc allele . | z1400 . | 15T.750 . | z4830 . | z3801 . |
| m294 | 11 (5/44) | 18 (8/44) | 26 (11/42) | 36 (15/41) |
| b16 | 0 (0/94) | 0 (0/96) | 0 (0/47) | 2 (1/66) |
| b229 | 0 (0/72) | 0 (0/113) | 3 (2/69) | 22 (15/69) |
| . | Percent recombination with cyc (No. rec./total) . | |||
|---|---|---|---|---|
| cyc allele . | z1400 . | 15T.750 . | z4830 . | z3801 . |
| m294 | 11 (5/44) | 18 (8/44) | 26 (11/42) | 36 (15/41) |
| b16 | 0 (0/94) | 0 (0/96) | 0 (0/47) | 2 (1/66) |
| b229 | 0 (0/72) | 0 (0/113) | 3 (2/69) | 22 (15/69) |
Genetic distance between cyc and LG 12 markers in cyc mutant mapping crosses
| . | Percent recombination with cyc (No. rec./total) . | |||
|---|---|---|---|---|
| cyc allele . | z1400 . | 15T.750 . | z4830 . | z3801 . |
| m294 | 11 (5/44) | 18 (8/44) | 26 (11/42) | 36 (15/41) |
| b16 | 0 (0/94) | 0 (0/96) | 0 (0/47) | 2 (1/66) |
| b229 | 0 (0/72) | 0 (0/113) | 3 (2/69) | 22 (15/69) |
| . | Percent recombination with cyc (No. rec./total) . | |||
|---|---|---|---|---|
| cyc allele . | z1400 . | 15T.750 . | z4830 . | z3801 . |
| m294 | 11 (5/44) | 18 (8/44) | 26 (11/42) | 36 (15/41) |
| b16 | 0 (0/94) | 0 (0/96) | 0 (0/47) | 2 (1/66) |
| b229 | 0 (0/72) | 0 (0/113) | 3 (2/69) | 22 (15/69) |
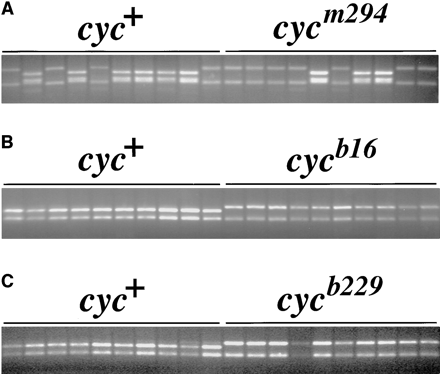
that both of these mutations suppress recombination between markers in the distal region of LG 12 (Figure 3 and Table 1). For example, 15T.750 frequently recombines with cycm294 but only rarely, if at all, recombines with cycb16 (Figure 3 and Table 1). Analysis of LG 12 markers in cycb16 mapping crosses revealed that recombination is suppressed in a region stretching from cyc to beyond marker z4830 (Table 1). Similarly, the cycb229 mutation suppresses recombination on LG 12 but over a smaller region than cycb16 (Figure 3 and Table 1). The cycb229 mutation failed to recombine with 15T.750 in 113 haploid individuals, whereas there were 8 recombinants for these loci among 44 individuals in cycm294 mapping crosses. More proximal markers, such as z3801, recombine frequently with cyc in cycb229 mapping crosses (Table 1), demarcating the region of recombination suppression by this mutation.
cycb213 involves a reciprocal translocation between LG 2 and LG 12: All three alleles discussed so far segregate according to Mendel among haploid offspring of heterozygous cyc+/− mothers; in these cases, about 50% of haploid embryos showed the mutant phenotype (Table 2). In contrast, the haploid offspring of cycb213/+ mothers display distinctly non-Mendelian segregation. Twenty-five percent (156/624) of the haploid progeny of cycb213/+ females had a cyclopic phenotype, whereas the Mendelian expectation is 50% (Table 2). Moreover, 25.8% (161/624) of the embryos in these clutches displayed
Markers near cyc fail to amplify from cycb16 and cycb229 genomic DNA. Individual haploid embryos obtained from mothers heterozygous for the cycb16 and cycb229 mutations were assayed with the SSLP markers z3467 and z1400. The z3467 fragment amplifies from the wild-type, but not the mutant, genomic DNA samples. Primers for both markers were included in the same PCR assays, so that z1400 serves an internal control for the mutant samples. Note that the two mutant chromosomes have different alleles of z1400, providing evidence that cycb229 is not an inadvertent reisolate of cycb16. Arrows indicate size standards of 100 nucleotides and 200 nucleotides in the marker lane (M).
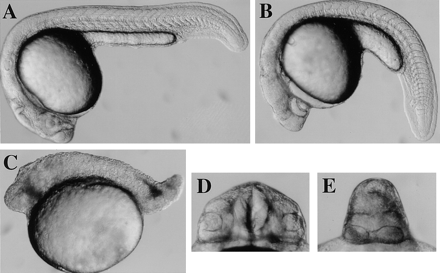
a characteristic necrotic mutant phenotype, which we have termed necb213 (Table 2 and Figure 4C; see materials and methods). This 2:1:1 segregation is characteristic of reciprocal translocations (Morgan et al. 1925),
Inheritance of cyclops alleles in haploid embryos
| cyc allele . | % wild-type (wild-type/total) . | % cyc− (cyc−/total) . | % necb213 (necb213/total) . |
|---|---|---|---|
| b16 | 51.6 (262/508) | 48.4 (246/508) | 0 |
| b229 | 48.4 (134/277) | 51.6 (143/277) | 0 |
| m294 | 47.1 (98/208) | 52.9 (110/208) | 0 |
| b213 | 49.2 (307/624) | 25.0 (156/624) | 25.8 (161/624) |
| cyc allele . | % wild-type (wild-type/total) . | % cyc− (cyc−/total) . | % necb213 (necb213/total) . |
|---|---|---|---|
| b16 | 51.6 (262/508) | 48.4 (246/508) | 0 |
| b229 | 48.4 (134/277) | 51.6 (143/277) | 0 |
| m294 | 47.1 (98/208) | 52.9 (110/208) | 0 |
| b213 | 49.2 (307/624) | 25.0 (156/624) | 25.8 (161/624) |
Inheritance of cyclops alleles in haploid embryos
| cyc allele . | % wild-type (wild-type/total) . | % cyc− (cyc−/total) . | % necb213 (necb213/total) . |
|---|---|---|---|
| b16 | 51.6 (262/508) | 48.4 (246/508) | 0 |
| b229 | 48.4 (134/277) | 51.6 (143/277) | 0 |
| m294 | 47.1 (98/208) | 52.9 (110/208) | 0 |
| b213 | 49.2 (307/624) | 25.0 (156/624) | 25.8 (161/624) |
| cyc allele . | % wild-type (wild-type/total) . | % cyc− (cyc−/total) . | % necb213 (necb213/total) . |
|---|---|---|---|
| b16 | 51.6 (262/508) | 48.4 (246/508) | 0 |
| b229 | 48.4 (134/277) | 51.6 (143/277) | 0 |
| m294 | 47.1 (98/208) | 52.9 (110/208) | 0 |
| b213 | 49.2 (307/624) | 25.0 (156/624) | 25.8 (161/624) |
Suppression of recombination by cycb16 and cycb229. An STS marker derived from RAPD 15T.750 was analyzed in haploid mapping crosses for (A) cycm294, (B) cycb16, and (C) cycb229. In each case, 10 wild-type and 10 mutant siblings are shown. The primers amplify allelic fragments of different sizes in all three crosses, but all fragments were nevertheless cleaved with the restriction enzyme Mnl I to generate fragments of optimal size for detecting the polymorphism with agarose gel electrophoresis.
suggesting that the b213 mutation may be a chromosomal rearrangement of this sort.
If b213 involves a reciprocal translocation with a breakpoint proximal to the cyc locus, then the cyclopic phenotype would result from the absence of the region of LG 12 that contains the cyc+ gene. As predicted by this model, markers from the distal region of LG 12 amplified from the genomic DNA of wild-type and necb213 haploid embryos, but not from their cycb213 siblings (Figure 5). The region of LG 12 proximal to z4830 is present in cycb213 individuals, whereas the markers in the segment distal to 15T.750 fail to amplify, indicating that the breakpoint on the rearranged chromosome lies 32 ± 6 cM proximal to cyc. This result indicates that the loss of cyc+ function in cycb213 mutants stems from the absence of cyc+ and other loci in the distal region of LG 12 rather than a breakpoint that disrupts the cyc+ gene.
If cycb213 involves a reciprocal translocation as suggested by the segregation data, and then the distal region of some linkage group other than LG 12 should be present in two copies in cycb213 embryos but absent in individuals with the necb213 phenotype. A systematic screen for markers absent from the DNA of necb213 mutants identified LG 2 as the translocation partner. Markers over a large segment of LG 2 (extending distally from twhh) did not amplify from necb213 embryo DNA (Figure 5 and data not shown). The proximal marker 12R.360 is present in these embryos, indicating
Wild-type (A, D) cycb213 (B, E) and necb213 (C) diploid embryos at 22 hours after fertilization. Side (A–C) and frontal (D, E) views are shown.
that the LG 2 breakpoint lies between 12R.360 and twhh.
Analysis of LG 2 and LG 12 markers in the haploid progeny of a b213/+ heterozygous female confirmed that these linkage groups are involved in a reciprocal translocation in the b213 mutation (Figures 6 and 7).
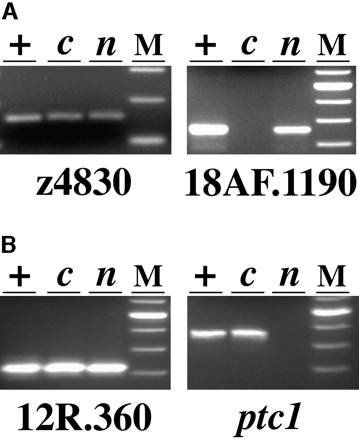
Analysis of LG 12 (A) and LG 2 (B) markers in haploid progeny of a b213/+ female. Pools of DNA from wild-type (lanes +), cycb213 (lanes c), and necb213 (lanes n) haploid embryos were tested with the SSLP z4830, STS markers derived from the RAPDs 18AF.1190 and 12R.360 and the gene patched1 (ptc1). z4830 and 12R.360 amplify from all three DNA samples, whereas 18AF.1190 fails to amplify from cycb213 DNA and ptc1 fails to amplify from necb213 DNA. Size standards (100-bp ladder, Life Technologies, Gaithersburg, MD) are shown in lanes M; fragment sizes are 160 bp for z4830, 360 bp for 18AF.1190, 330 bp for 12R.360, and 480 bp for ptc1.
The 350-bp allele of the LG 12 marker 15T.750 cosegregates with the 330-bp allele of the LG 2 marker 12R.360 (arrowheads in Figure 6), demonstrating that these loci lie together on a rearranged chromosome, T(LG2; LG12)b213, 2P12D, comprised of the centromeric region of LG 2 and the distal region of LG 12 (Figure 7). In contrast, the 320-bp allele of the LG 12 marker 15T.750 (arrow in Figure 6) segregates independepently of the 350-bp allele of the same locus in the b213 mapping panel, as would be expected for markers on different chromosomes, but not for alleles of the same locus. The wild-type individuals inherited one (never zero or two) 15T.750 allele, which is expected for allelic segregation in haploid embryos. However, the cycb213 animals inherited neither 15T.750 allele and the necb213 animals inherited both, as expected for loci on different chromosomes. In similar experiments (summarized in Figure 7A), markers from the proximal region of LG 12, including z4830, cosegregate with markers from the distal region of LG 2, including eng3, confirming that the b213 mutation involves a reciprocally rearranged chromosome, termed T(LG2;LG12)b213, 12P2D, comprised of the centromeric region of LG 12 and the distal region of LG 2 (Figure 7). The combined results demonstrate that the diploid mother of the b213 haploid mapping family was a balanced translocation heterozygote, possessing a normal order LG 2, a normal order LG 12, and two reciprocally rearranged LG 2-LG 12 chromosomes (Figure 7). Analysis of the inheritance of the rearranged chromosomes and the resulting phenotypes indicates that the wild-type embryos are euploid, inheriting both or neither rearranged chromosomes (schematized as wild type* and wild type, respectively, in Figure 7B) and that the cycb213 and necb213 embryos are aneuploid, having a deletion of either distal LG 12 or
LG 2 and LG 12 markers cosegregate in a b213 mapping cross. Individual haploid progeny of a DAR x b213 F1 female were analyzed with STS markers derived from the RAPDs 12R.360 (LG 2) and 15T.750 (LG 12). Both markers amplify allelic fragments of different sizes. The 12R.360 alleles are 360 bp and 330 bp (arrowhead). The 15T.750 fragments were cleaved with the restriction enzyme Mnl I to generate fragments of optimal size for detecting the polymorphism with agarose gel electrophoresis. The allele-specific fragments are 320 bp (arrow) and 350 bp (arrowhead); the common fragment is 270 bp. necb213 haploids inherit both alleles of 15T.750, whereas cycb213 embryos inherit neither allele; the faint bands in the cycb213 samples are nonspecific amplification products.
distal LG 2 and a duplication of the reciprocal fragment.
DISCUSSION
Recombination suppression by cycb16 and cycb229: The results show that the gamma-ray induced cycb16 and cycb229 mutations contain deletions in the distal region of LG 12. Interestingly, both mutations suppress recombination between genetic markers spanning more than 30 cM of LG 12, including a region far removed from the deletion. It is possible that both cycb16 and cycb229 are complex rearrangements, with other aberrations accounting for the recombination suppression. For example, each allele could bear an inversion for a segment of LG 12 in addition to a deletion in the vicinity of cyc.
An intriguing alternative is that the cycb16 and cycb229 deletions affect a chromosomal site that is required for recombination of markers at a distance. Precedent for this derives from work on meiotic pairing and crossing over in Caenorhabditis elegans (McKim et al. 1988; Villeneuve 1994; Wicky and Rose 1996). In this species, analysis of the effects of chromosomal rearrangements on recombination has led to the proposal that a region near one end of each chromosome functions as a cis-acting pairing site required for homolog recognition, recombination, and disjunction in meiosis. The parallel between the present study and the C. elegans pairing sites is striking, as in both cases removal of a region near one end of a chromosome reduces recombination between markers elsewhere on the chromosome. The cycb16 and cycb229 mutations suppress recombination to different extents (Table 1), suggesting that if these effects are caused by deletion of a site required for crossing over, then activity of the site is distributed over a region that is differentially affected by the two mutations. Whatever the mechanism of recombination suppression, the cycb16 and cycb229 mutations may be useful for balancing other lethal mutations located in the distal region of LG 12.
Translocations in zebrafish: We have shown that the T(LG2;LG12)b213 mutation is a reciprocal translocation between LG 12, where cyc resides, and LG 2. This conclusion
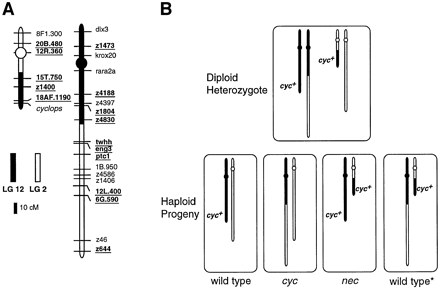
Reciprocally rearranged chromosomes in T(LG2;LG12)b213. (A) The positions of the breakpoints were determined by analyzing LG 2 and LG 12 markers (underlined) in pooled or individual DNA samples from the b213 haploid mapping family shown in Figure 6. Marker distances from previous wild-type maps (Postlethwait et al. 1994; Johnson et al. 1996) were used to determine the lengths of segments involved in the rearrangements. Markers not underlined were not analyzed in the b213 mapping panel; their positions are inferred from the previous maps cited above. Centromere positions (circles) have been reported (Johnson et al. 1996). (B) Schematic showing haploid progeny derived from a balanced translocation heterozygote. Half of the haploid embryos inherit the rearranged LG 12 (proximal LG 12-distal LG 2) that lacks cyc+. Half of these individuals (wild type*) also inherit the rearranged LG 2 (proximal LG 2-distal LG 12), and so they have a cyc+ gene and are euploid. These animals have a wild-type phenotype, indicating that neither breakpoint results in a phenotype morphologically detectable in haploid embryos. The other animals inheriting the rearranged LG 12 (cyc) also inherit a normal order LG 2, which does not carry the cyc+ gene. The necb213 phenotype occurs in embryos inheriting a rearranged LG 2 and a normal order LG 12.
derives from the segregation of the cycb213 and necb213 mutant phenotypes and also from demonstration of rearranged T(LG2;LG12)b213, 12P2D and T(LG2; LG12)b213, 2P12D chromosomes by analysis of genetic markers. The cycb213 and necb213 mutant phenotypes result from aneuploidy for the distal regions of LG 12 and LG 2 that measure ~33 ± 6 cM and ~125 ± 20 cM, respectively. Thus cycb213 mutants are deficient for approximately 1% of the 3000 cM genome (Postlethwait et al. 1994), a region that surely contains many genes. Despite this, the cycb213 mutant phenotype during embryogenesis is similar to that caused by the ENU-induced allele cycm294 (Schier et al. 1996), and homozygotes for both cyc alleles survive until just before the end of embryogenesis. This suggests that cyc function is essential earlier in development of the zygote than other genes in the region. In contrast, the phenotype of necb213 mutants is quite severe, and these animals do not typically survive beyond 30 hr. The large size of the deficient segment in necb213 mutants and the nature of the phenotype suggest that multiple genes with early, essential functions are deleted in these animals. Although these aneuploid phenotypes cannot be attributed to the functions of individual genes in the deficient segments, these rearrangements are useful in concert with single-gene mutations in the definition of the amorphic phenotypes of genes in the translocated segments.
Translocations have important applications in mapping cloned genes and mutations. For example, patched1 (ptc1; Figure 5) and other genes (Figure 7 and data not shown) were assigned to the region of LG 2 deleted from the T(LG2;LG12)b213, 2P12D chromosome by virtue of their failure to amplify from genomic DNA of the aneuploid animals lacking this segment. Thus a panel of translocations covering the genome would be useful for mapping cloned genes without the need to identify linked polymorphisms. The resolution of translocation panel mapping would be significantly less than traditional genetic mapping (Postlethwait et al. 1994) and radiation hybrid approaches (Cox et al. 1990), and perhaps comparable to mapping with somatic cell hybrids, which also assigns genes to chromosomes or large chromosomal segments (Ekker et al. 1996). However, a translocation panel has the important additional advantage that it can be applied to the mapping of mutations. By crossing heterozygotes to a panel of translocation stocks, a mutation can be mapped to a deleted chromosomal segment by failure of complementation.
A number of other gamma-ray induced rearrangements have been described previously (Fritz et al. 1996; Fisher et al. 1997), and these mutations together with the T(LG2;LG12)b213 translocation collectively cover more than 500 cM of the genetic map, about 17% of the total. For comparison, 10–15% of the mouse genome is covered by large deletions (Brown and Peters 1996). Thus significant progress toward assembling a translocation panel has already been achieved. Methods that couple the use of haploid embryos for simplifying segregation analysis and the use of PCR (Fritz et al. 1996) for targeting specific regions of the genome marked by cloned genes or SSLPs should enable the construction of an expanded panel of translocations.
Acknowledgement
We thank Scott Hawley, Ruth Lehmann, Alexander Schier, and the members of our laboratories for helpful discussions; Wolfgang Driever for support during the screen that identified the cycm294 allele; and Tim Carl for technical assistance. This work was supported by grants R01 RR12349 (to W.S.T.), P01 HD22486 (to J.H.P. and C.B.K.), R01 RR10715 (to J.H.P.), and R01 HD29761 (to W. Driever) from the National Institutes of Health.
Footnotes
Communicating editor: N. A. Jenkins
LITERATURE CITED
Author notes
Present address: Max-Planck Institut für Entwicklungsbiologie, Spemannstrasse 35/I, 72076 Tübingen, Germany.